Linking Karyotypes with DNA Barcodes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Distributional Study of the Butterflies of the Sierra De Tuxtla in Veracruz, Mexico. Gary Noel Ross Louisiana State University and Agricultural & Mechanical College
Louisiana State University LSU Digital Commons LSU Historical Dissertations and Theses Graduate School 1967 A Distributional Study of the Butterflies of the Sierra De Tuxtla in Veracruz, Mexico. Gary Noel Ross Louisiana State University and Agricultural & Mechanical College Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_disstheses Recommended Citation Ross, Gary Noel, "A Distributional Study of the Butterflies of the Sierra De Tuxtla in Veracruz, Mexico." (1967). LSU Historical Dissertations and Theses. 1315. https://digitalcommons.lsu.edu/gradschool_disstheses/1315 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Historical Dissertations and Theses by an authorized administrator of LSU Digital Commons. For more information, please contact [email protected]. This dissertation has been microfilmed exactly as received 67-14,010 ROSS, Gary Noel, 1940- A DISTRIBUTIONAL STUDY OF THE BUTTERFLIES OF THE SIERRA DE TUXTLA IN VERACRUZ, MEXICO. Louisiana State University and Agricultural and Mechanical CoUege, Ph.D., 1967 Entomology University Microfilms, Inc., Ann Arbor, Michigan A DISTRIBUTIONAL STUDY OF THE BUTTERFLIES OF THE SIERRA DE TUXTLA IN VERACRUZ, MEXICO A D issertation Submitted to the Graduate Faculty of the Louisiana State University and A gricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Department of Entomology by Gary Noel Ross M.S., Louisiana State University, 196*+ May, 1967 FRONTISPIECE Section of the south wall of the crater of Volcan Santa Marta. May 1965, 5,100 feet. ACKNOWLEDGMENTS Many persons have contributed to and assisted me in the prep aration of this dissertation and I wish to express my sincerest ap preciation to them all. -

INSECTA: LEPIDOPTERA) DE GUATEMALA CON UNA RESEÑA HISTÓRICA Towards a Synthesis of the Papilionoidea (Insecta: Lepidoptera) from Guatemala with a Historical Sketch
ZOOLOGÍA-TAXONOMÍA www.unal.edu.co/icn/publicaciones/caldasia.htm Caldasia 31(2):407-440. 2009 HACIA UNA SÍNTESIS DE LOS PAPILIONOIDEA (INSECTA: LEPIDOPTERA) DE GUATEMALA CON UNA RESEÑA HISTÓRICA Towards a synthesis of the Papilionoidea (Insecta: Lepidoptera) from Guatemala with a historical sketch JOSÉ LUIS SALINAS-GUTIÉRREZ El Colegio de la Frontera Sur (ECOSUR). Unidad Chetumal. Av. Centenario km. 5.5, A. P. 424, C. P. 77900. Chetumal, Quintana Roo, México, México. [email protected] CLAUDIO MÉNDEZ Escuela de Biología, Universidad de San Carlos, Ciudad Universitaria, Campus Central USAC, Zona 12. Guatemala, Guatemala. [email protected] MERCEDES BARRIOS Centro de Estudios Conservacionistas (CECON), Universidad de San Carlos, Avenida La Reforma 0-53, Zona 10, Guatemala, Guatemala. [email protected] CARMEN POZO El Colegio de la Frontera Sur (ECOSUR). Unidad Chetumal. Av. Centenario km. 5.5, A. P. 424, C. P. 77900. Chetumal, Quintana Roo, México, México. [email protected] JORGE LLORENTE-BOUSQUETS Museo de Zoología, Facultad de Ciencias, UNAM. Apartado Postal 70-399, México D.F. 04510; México. [email protected]. Autor responsable. RESUMEN La riqueza biológica de Mesoamérica es enorme. Dentro de esta gran área geográfi ca se encuentran algunos de los ecosistemas más diversos del planeta (selvas tropicales), así como varios de los principales centros de endemismo en el mundo (bosques nublados). Países como Guatemala, en esta gran área biogeográfi ca, tiene grandes zonas de bosque húmedo tropical y bosque mesófi lo, por esta razón es muy importante para analizar la diversidad en la región. Lamentablemente, la fauna de mariposas de Guatemala es poco conocida y por lo tanto, es necesario llevar a cabo un estudio y análisis de la composición y la diversidad de las mariposas (Lepidoptera: Papilionoidea) en Guatemala. -

Archiv Für Naturgeschichte
© Biodiversity Heritage Library, http://www.biodiversitylibrary.org/; www.zobodat.at Lepidoptera für 1903. Bearbeitet von Dr. Robert Lucas in Rixdorf bei Berlin. A. Publikationen (Autoren alphabetisch) mit Referaten. Adkin, Robert. Pyrameis cardui, Plusia gamma and Nemophila noc- tuella. The Entomologist, vol. 36. p. 274—276. Agassiz, G. Etüde sur la coloration des ailes des papillons. Lausanne, H. Vallotton u. Toso. 8 °. 31 p. von Aigner-Abafi, A. (1). Variabilität zweier Lepidopterenarten. Verhandlgn. zool.-bot. Ges. Wien, 53. Bd. p. 162—165. I. Argynnis Paphia L. ; IL Larentia bilineata L. — (2). Protoparce convolvuli. Entom. Zeitschr. Guben. 17. Jahrg. p. 22. — (3). Über Mimikry. Gaea. 39. Jhg. p. 166—170, 233—237. — (4). A mimicryröl. Rov. Lapok, vol. X, p. 28—34, 45—53 — (5). A Mimicry. Allat. Kozl. 1902, p. 117—126. — (6). (Über Mimikry). Allgem. Zeitschr. f. Entom. 7. Bd. (Schluß p. 405—409). Über Falterarten, welche auch gesondert von ihrer Umgebung, in ruhendem Zustande eine eigentümliche, das Auge täuschende Form annehmen (Lasiocampa quercifolia [dürres Blatt], Phalera bucephala [zerbrochenes Ästchen], Calocampa exoleta [Stück morschen Holzes]. — [Stabheuschrecke, Acanthoderus]. Raupen, die Meister der Mimikry sind. Nachahmung anderer Tiere. Die Mimik ist in vielen Fällen zwecklos. — Die wenn auch recht geistreichen Mimikry-Theorien sind doch vielleicht nur ein müßiges Spiel der Phantasie. Aitken u. Comber, E. A list of the butterflies of the Konkau. Journ. Bombay Soc. vol. XV. p. 42—55, Suppl. p. 356. Albisson, J. Notes biologiques pour servir ä l'histoire naturelle du Charaxes jasius. Bull. Soc. Etud. Sc. nat. Nimes. T. 30. p. 77—82. Annandale u. Robinson. Siehe unter S w i n h o e. -

With Descriptions of Nine New Species
European Journal of Taxonomy 551: 1–67 ISSN 2118-9773 https://doi.org/10.5852/ejt.2019.551 www.europeanjournaloftaxonomy.eu 2019 · Nakahara S. et al. This work is licensed under a Creative Commons Attribution License (CC BY 4.0). Monograph urn:lsid:zoobank.org:pub:C3C851C3-0F12-412C-A15B-56F0F263CD00 Revision of the poorly known Neotropical butterfly genus Zischkaia Forster, 1964 (Lepidoptera, Nymphalidae, Satyrinae), with descriptions of nine new species Shinichi NAKAHARA 1,*, Thamara ZACCA 2, Fernando M.S. DIAS 3, Diego R. DOLIBAINA 4, Lei XIAO 5, Marianne ESPELAND 6, Mirna M. CASAGRANDE 7, Olaf H.H. MIELKE 8, Gerardo LAMAS 9, Blanca HUERTAS 10, Kaylin KLECKNER 11 & Keith R. WILLMOTT 12 1,5,11,12 McGuire Center for Lepidoptera and Biodiversity, Florida Museum of Natural History, University of Florida, Gainesville, Florida 32611, USA. 1,11 Department of Entomology and Nematology, University of Florida, Gainesville, Florida 32611, USA. 2 Departamento de Biologia Animal and Museu de Zoologia, Instituto de Biologia, Universidade Estadual de Campinas, Campinas, São Paulo, Brazil. 3,4,7,8 Laboratório de Estudos de Lepidoptera Neotropical, Departamento de Zoologia, Universidade Federal do Paraná, Curitiba, Paraná, Brazil. 6 Arthropoda Department, Zoological Research Museum Alexander Koenig, Adenauer Allee 160, 53113 Bonn, Germany. 1,9 Departamento de Entomología, Museo de Historia Natural, Universidad Nacional Mayor de San Marcos, Lima, Peru. 10 Life Sciences Department, Natural History Museum, London, UK. * Corresponding author: [email protected] -

The Brain of a Nocturnal Migratory Insect, the Australian Bogong Moth
bioRxiv preprint doi: https://doi.org/10.1101/810895; this version posted January 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The brain of a nocturnal migratory insect, the Australian Bogong moth Authors: Andrea Adden1, Sara Wibrand1, Keram Pfeiffer2, Eric Warrant1, Stanley Heinze1,3 1 Lund Vision Group, Lund University, Sweden 2 University of Würzburg, Germany 3 NanoLund, Lund University, Sweden Correspondence: [email protected] Abstract Every year, millions of Australian Bogong moths (Agrotis infusa) complete an astonishing journey: in spring, they migrate over 1000 km from their breeding grounds to the alpine regions of the Snowy Mountains, where they endure the hot summer in the cool climate of alpine caves. In autumn, the moths return to their breeding grounds, where they mate, lay eggs and die. These moths can use visual cues in combination with the geomagnetic field to guide their flight, but how these cues are processed and integrated in the brain to drive migratory behavior is unknown. To generate an access point for functional studies, we provide a detailed description of the Bogong moth’s brain. Based on immunohistochemical stainings against synapsin and serotonin (5HT), we describe the overall layout as well as the fine structure of all major neuropils, including the regions that have previously been implicated in compass-based navigation. The resulting average brain atlas consists of 3D reconstructions of 25 separate neuropils, comprising the most detailed account of a moth brain to date. -
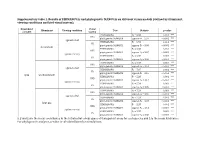
Supplementary Table 1. Results of Permanovas and Phylogenetic Manovas on Different Vision Models (Defined by Illuminant, Viewing Conditions and Bird Visual System)
Supplementary table 1. Results of PERMANOVAs and phylogenetic MANOVAs on different vision models (defined by illuminant, viewing conditions and bird visual system). Dependent Visual Illuminant Viewing condition Test Statistic p-value variable system PERMANOVA F9 = 6.88 0.001 *** UVS phylogenetic MANOVA approx-F9 = 2.97 < 0.001 *** against a leaf PERMANOVA F9 = 6.93 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.05 < 0.001 *** forest shade PERMANOVA F9 = 5.38 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.07 < 0.001 *** against the sky PERMANOVA F9 = 5.38 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.36 < 0.001 *** PERMANOVA F9 = 7.04 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.01 < 0.001 *** against a leaf PERMANOVA F9 = 7.07 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.10 < 0.001 *** xyzL woodland shade PERMANOVA F9 = 5.33 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.12 < 0.001 *** against the sky PERMANOVA F9 = 5.34 0.002 ** VS phylogenetic MANOVA approx-F9 = 3.39 < 0.001 *** PERMANOVA F9 = 7.24 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.00 < 0.001 *** against a leaf PERMANOVA F9 = 7.24 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.07 < 0.001 *** large gap PERMANOVA F9 = 5.37 0.001 *** UVS phylogenetic MANOVA approx-F9 = 3.14 < 0.001 *** against the sky PERMANOVA F9 = 5.37 0.001 *** VS phylogenetic MANOVA approx-F9 = 3.38 < 0.001 *** x, y and z are the mean coordinates in the tetrahedral colour space of transparent areas for each species and L is the mean luminance. -

Re-Emergence and Diversification of a Specialised Antennal Lobe Morphology in Ithomiine Butterflies
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.13.336206; this version posted October 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Re-emergence and diversification of a specialised antennal lobe morphology 2 in ithomiine butterflies 3 4 Authors: 5 Billy J Morris1*, Antoine Couto1,2, Asli Aydin3, Stephen H Montgomery2*. 6 7 Affiliations: 1 8 Dept. of Zoology, University of Cambridge, Downing Street, Cambridge, CB2 3EJ 9 2 School of Biological Sciences, University of Bristol, 24 Tyndall Avenue, Bristol, BS8 1TQ 10 3 School of Medicine, Koc University, Rumelifeneri Yolu 34450 Sarıyer / Istanbul, Turkey 11 12 * corresponding authors: 13 BJM: [email protected] 14 SHM: [email protected] 15 16 Abstract 17 How an organism’s sensory system functions is central to how it navigates its environment and 18 meets the behavioural challenges associated with survival and reproduction. Comparing sensory 19 systems across species can reveal how facets of behaviour and ecology promote adaptive shifts 20 in the relative importance of certain environmental cues. The insect olfactory system is prominent 21 model for investigating how ecological factors impact sensory reception and processing. Notably 22 work in Lepidoptera led to the discovery of vastly expanded structures, termed a macroglomerular 23 complex (MGC), within the primary olfactory processing centre. These structures typically process 24 pheromonal cues and provide a classic example of how variation in size can influence the 25 functional processing of sensory cues. -

Rev Iss Web Mec 13773 25-22 5765..5784
Molecular Ecology (2016) 25, 5765–5784 doi: 10.1111/mec.13773 Into the Andes: multiple independent colonizations drive montane diversity in the Neotropical clearwing butterflies Godyridina NICOLAS CHAZOT,*† KEITH R. WILLMOTT,‡ FABIEN L. CONDAMINE,§¶ DONNA LISA DE-SILVA,* ANDREV.L.FREITAS,**GERARDOLAMAS,†† HELENE MORLON,‡‡ CARLOS E. GIRALDO,§§ CHRIS D. JIGGINS,¶¶ MATHIEU JORON,*** JAMES MALLET,††† SANDRA URIBE‡‡‡ and MARIANNE ELIAS* *Institut de Systematique, Evolution, Biodiversite, ISYEB – UMR 7205 – CNRS MNHN UPMC EPHE, Museum national d’Histoire naturelle, Sorbonne Universites, 57 rue Cuvier CP50, F-75005 Paris, France, †Department of Biology, University of Lund, 223 62 Lund, Sweden, ‡McGuire Center for Lepidoptera and Biodiversity, Florida Museum of Natural History, University of Florida, Gainesville, FL 32611, USA, §CNRS, UMR 5554 Institut des Sciences de l’Evolution (Universite de Montpellier), Place Eugene Bataillon, 34095 Montpellier, France, ¶Department of Biological Sciences, University of Alberta, T6G 2E9 Edmonton, AB, Canada, **Departamento de Zoologia and Museu de Zoologia, Instituto de Biologia, Universidade Estadual de Campinas, Campinas, S~ao Paulo, Brazil, ††Museo de Historia Natural, Universidad Nacional de San Marcos, Lima, Peru, ‡‡IBENS, Ecole Normale Superieure, UMR 8197 CNRS, Paris, France, §§Grupo de Investigacion de Sanidad Vegetal, Universidad Catolica de Oriente, Rionegro, Antioquia, Colombia, ¶¶Department of Zoology, University of Cambridge, Cambridge, UK, ***Centre d’Ecologie Fonctionnelle et Evolutive, CEFE, UMR 5175 CNRS – EPHE – Universite de Montpellier – Universite Paul Valery Montpellier, 34293 Montpellier 5, France, †††Department of Organismic and Evolutionary Biology, Harvard University, Cambridge, MA 02138, USA, ‡‡‡Universidad Nacional de Colombia, sede Medellın, Medellın, Colombia Abstract Understanding why species richness peaks along the Andes is a fundamental question in the study of Neotropical biodiversity. -

Panama's Brilliant Butterflies December 3-15, 2017
Panama's Brilliant Butterflies December 3-15, 2017 Guides: Faustino "Tino" Sanchez Daily Itinerary : Linda Harrison Day 1 - December 3rd; Arrival, Canopy Lodge gardens Day 2 - December 4th; Cara Iguana am & Las Mozas pm Day 3 - December 5th; Las Minas/Cara Iguana am & Finca Macarena/Las Minas pm TOTAL SPECIES: 301 (including 5?) Day 4 - December 6th; Altos del Maria & Canopy Lodge gardens Day 5 - December 7th; Canopy Lodge am & Canopy Tower/Semaphore Hill pm Main Tour: 235 Day 6 - December 8th; Ammo Ponds & Pipeline Road Day 7 - December 9th; Metropolitan Park, Canopy Tower & Summit Ponds Camp: 144 Day 8 - December 10th; Canopy Tower Canopy Camp Extension : Blue=species added Main Tour: 6 Day 8 - December 10th; Bayano Lake, Torti & Canopy Camp grounds Orange=species added Camp Tour: 4 Day 9 - December 11th; Canopy Camp am & Pan American Hwy. pm Day 10 - December 12th; El Salto Road am & Pan American Hwy. pm Day 11 - December 13th; El Salto Road am & Lagos Blancas pm There are a few trip photos at end! Day 12 - December 14th; Aligondi am & Sansom pm Day 13 - December 15th; San Francisco Reserve am 3rd 4th 5th 6th 7th 8th 9th 10th 11th 12th 13th 14th 15th PAPILIONIDAE SWALLOWTAILS Papilioninae Swallowtails & Cattlehearts Neographium agesilaus Short-lined Kite-Swallowtail X Battus polydamas Polydamas Swallowtail X Battus ingenuus Dyar's Swallowtail X Parides childrenae childrenae Green-celled Cattleheart X X X X X X Parides sesostris tarquinius Emerald-patched Cattleheart X X X XCC X X Heraclides anchisiades Ruby-spotted Swallowtail X Heraclides -

Molecular Phylogenetics of the Neotropical Butterfly Subtribe Oleriina
Molecular Phylogenetics and Evolution 55 (2010) 1032–1041 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Molecular phylogenetics of the neotropical butterfly subtribe Oleriina (Nymphalidae: Danainae: Ithomiini) Donna Lisa de-Silva a,*, Julia J. Day a, Marianne Elias b,c, Keith Willmott d, Alaine Whinnett a, James Mallet a a Department of Genetics, Evolution and Environment, University College London, Wolfson House, 4 Stephenson Way, London NW1 2HE, UK b Imperial College London, Silwood Park, Buckhurst Road, Ascot, Berkshire SL5 7PY, UK c CNRS, UMR 7205, Muséum National d’Histoire Naturelle, 45 Rue Buffon, CP50, 75005 Paris, France d McGuire Center for Lepidoptera, Florida Museum of Natural History, University of Florida, P.O. Box 112710, Gainesville, FL 32611-2710, USA article info abstract Article history: The Oleriina is one of the most speciose subtribes of the neotropical nymphalid butterfly tribe Ithomiini. Received 9 September 2009 They are widely distributed across the Andes and Amazonian lowlands and like other ithomiines they are Revised 22 December 2009 involved in complex mimicry rings. This subtribe is of particular interest because it contains the most Accepted 9 January 2010 diverse ithomiine genus, Oleria, as well as two genera, Megoleria and Hyposcada, that feed on hostplants Available online 15 January 2010 not utilized elsewhere in the tribe. Here we present the first comprehensive species-level phylogeny for the Oleriina, representing 83% of recognised species in the group, and based on 6698 bp from eight mito- Keywords: chondrial (mt) and nuclear (nc) genes. Topologies are largely congruent for ncDNA and the concatenated Lepidoptera dataset and the genera Oleria, Hyposcada and Megoleria are recovered and well-supported, although Speciation Phylogeny strongly discordant genealogy between mtDNA and ncDNA suggest possible introgression among Hypos- Hybridization cada and Megoleria. -

Effects of Land Use on Butterfly (Lepidoptera: Nymphalidae) Abundance and Diversity in the Tropical Coastal Regions of Guyana and Australia
ResearchOnline@JCU This file is part of the following work: Sambhu, Hemchandranauth (2018) Effects of land use on butterfly (Lepidoptera: Nymphalidae) abundance and diversity in the tropical coastal regions of Guyana and Australia. PhD Thesis, James Cook University. Access to this file is available from: https://doi.org/10.25903/5bd8e93df512e Copyright © 2018 Hemchandranauth Sambhu The author has certified to JCU that they have made a reasonable effort to gain permission and acknowledge the owners of any third party copyright material included in this document. If you believe that this is not the case, please email [email protected] EFFECTS OF LAND USE ON BUTTERFLY (LEPIDOPTERA: NYMPHALIDAE) ABUNDANCE AND DIVERSITY IN THE TROPICAL COASTAL REGIONS OF GUYANA AND AUSTRALIA _____________________________________________ By: Hemchandranauth Sambhu B.Sc. (Biology), University of Guyana, Guyana M.Sc. (Res: Plant and Environmental Sciences), University of Warwick, United Kingdom A thesis Prepared for the College of Science and Engineering, in partial fulfillment of the requirements for the degree of Doctor of Philosophy James Cook University February, 2018 DEDICATION ________________________________________________________ I dedicate this thesis to my wife, Alliea, and to our little girl who is yet to make her first appearance in this world. i ACKNOWLEDGEMENTS ________________________________________________________ I would like to thank the Australian Government through their Department of Foreign Affairs and Trade for graciously offering me a scholarship (Australia Aid Award – AusAid) to study in Australia. From the time of my departure from my home country in 2014, Alex Salvador, Katherine Elliott and other members of the AusAid team have always ensured that the highest quality of care was extended to me as a foreign student in a distant land. -

Variable Selection and the Coexistence of Multiple Mimetic Forms of the Butterfly Heliconius Numata
Evolutionary Ecology 13: 721±754, 1999. Ó 2001 Kluwer Academic Publishers. Printed in the Netherlands. Research paper Variable selection and the coexistence of multiple mimetic forms of the butter¯y Heliconius numata MATHIEU JORON1*, IAN R. WYNNE2, GERARDO LAMAS3 and JAMES MALLET2 1Institut des Sciences de l'Evolution, Universite de MontpellierII, Place E. Bataillon, F-34095 Montpellier, France; 2Department of Biology, University College London, 4, Stephenson Way, London NW1 2HE, UK; 3Departamento de EntomologõÂa, Museo de Historia Natural, Universidad Nacional Mayor de San Marcos, Apartado 14-0434, Lima-14, Peru (*author for correspondence, fax: +31 (0) 71-527-4900; email: [email protected] Inst. Evol. Ecol. Sciences, Univ. Leiden, PO BOX 9516, 2300 RA Leiden, The Netherlands) Received 20 June 2000; accepted 11 December 2000 Co-ordinating editor: C. Rowe Abstract. Polymorphism in aposematic animals and coexistence of multiple mimicry rings within a habitat are not predicted by classical MuÈ llerian mimicry. The butter¯y Heliconius numata Cramer (Lepidoptera: Nymphalidae; Heliconiinae) is both polymorphic and aposematic. The polymor- phism is due to variation at a single locus (or `supergene') which determines colour patterns involved in MuÈ llerian mimicry. We sampled 11 sites in a small area (approx. 60 ´ 30 km) of North- eastern Peru for H. numata and its co-mimics in the genus Melinaea and Athyrtis (Ithomiinae), and examined the role of temporal and spatial heterogeneity in the maintenance of polymorphism. Colour-patterns of Melinaea communities, which constitute the likely `mimetic environment' for H. numata, are dierentiated on a more local scale than morphs of H. numata, but the latter do show a strong and signi®cant response to local selection for colour-pattern.