Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Transcriptome Response of High- and Low-Light-Adapted Prochlorococcus Strains to Changing Iron Availability
Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Thompson, Anne W et al. “Transcriptome Response of High- and Low-light-adapted Prochlorococcus Strains to Changing Iron Availability.” ISME Journal (2011), 1-15. As Published http://dx.doi.org/10.1038/ismej.2011.49 Publisher Nature Publishing Group Version Author's final manuscript Citable link http://hdl.handle.net/1721.1/64705 Terms of Use Creative Commons Attribution-Noncommercial-Share Alike 3.0 Detailed Terms http://creativecommons.org/licenses/by-nc-sa/3.0/ Title: Transcriptome response of high- and low-light adapted Prochlorococcus strains to changing iron availability Running title: Prochlorococcus response to iron stress 5 Contributors: Anne W. Thompson1, Katherine Huang1, Mak A. Saito* 2, Sallie W. Chisholm* 1, 3 10 1 MIT Department of Civil and Environmental Engineering 2 Woods Hole Oceanographic Institution – Department of Marine Chemistry and Geochemistry 3 MIT Department of Biology 15 * To whom correspondence should be addressed: E-mail: [email protected] and [email protected] Subject Category: Microbial population and community ecology 20 Abstract Prochlorococcus contributes significantly to ocean primary productivity. The link between primary productivity and iron in specific ocean regions is well established and iron-limitation of Prochlorococcus cell division rates in these regions has been 25 demonstrated. However, the extent of ecotypic variation in iron metabolism among Prochlorococcus and the molecular basis for differences is not understood. Here, we examine the growth and transcriptional response of Prochlorococcus strains, MED4 and MIT9313, to changing iron concentrations. -

View of HER2: Human Epidermal Growth Factor Receptor 2; TNBC: Triple-Negative Breast Resistance to Systemic Therapy in Patients with Breast Cancer
Wen et al. Cancer Cell Int (2018) 18:128 https://doi.org/10.1186/s12935-018-0625-9 Cancer Cell International PRIMARY RESEARCH Open Access Sulbactam‑enhanced cytotoxicity of doxorubicin in breast cancer cells Shao‑hsuan Wen1†, Shey‑chiang Su2†, Bo‑huang Liou3, Cheng‑hao Lin1 and Kuan‑rong Lee1* Abstract Background: Multidrug resistance (MDR) is a major obstacle in breast cancer treatment. The predominant mecha‑ nism underlying MDR is an increase in the activity of adenosine triphosphate (ATP)-dependent drug efux trans‑ porters. Sulbactam, a β-lactamase inhibitor, is generally combined with β-lactam antibiotics for treating bacterial infections. However, sulbactam alone can be used to treat Acinetobacter baumannii infections because it inhibits the expression of ATP-binding cassette (ABC) transporter proteins. This is the frst study to report the efects of sulbactam on mammalian cells. Methods: We used the breast cancer cell lines as a model system to determine whether sulbactam afects cancer cells. The cell viabilities in the present of doxorubicin with or without sulbactam were measured by MTT assay. Protein identities and the changes in protein expression levels in the cells after sulbactam and doxorubicin treatment were determined using LC–MS/MS. Real-time reverse transcription polymerase chain reaction (real-time RT-PCR) was used to analyze the change in mRNA expression levels of ABC transporters after treatment of doxorubicin with or without sulbactam. The efux of doxorubicin was measures by the doxorubicin efux assay. Results: MTT assay revealed that sulbactam enhanced the cytotoxicity of doxorubicin in breast cancer cells. The results of proteomics showed that ABC transporter proteins and proteins associated with the process of transcription and initiation of translation were reduced. -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

Transcriptomic Analysis of the Aquaporin (AQP) Gene Family
Pancreatology 19 (2019) 436e442 Contents lists available at ScienceDirect Pancreatology journal homepage: www.elsevier.com/locate/pan Transcriptomic analysis of the Aquaporin (AQP) gene family interactome identifies a molecular panel of four prognostic markers in patients with pancreatic ductal adenocarcinoma Dimitrios E. Magouliotis a, b, Vasiliki S. Tasiopoulou c, Konstantinos Dimas d, * Nikos Sakellaridis d, Konstantina A. Svokos e, Alexis A. Svokos f, Dimitris Zacharoulis b, a Division of Surgery and Interventional Science, Faculty of Medical Sciences, UCL, London, UK b Department of Surgery, University of Thessaly, Biopolis, Larissa, Greece c Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece d Department of Pharmacology, Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece e The Warren Alpert Medical School of Brown University, Providence, RI, USA f Riverside Regional Medical Center, Newport News, VA, USA article info abstract Article history: Background: This study aimed to assess the differential gene expression of aquaporin (AQP) gene family Received 14 October 2018 interactome in pancreatic ductal adenocarcinoma (PDAC) using data mining techniques to identify novel Received in revised form candidate genes intervening in the pathogenicity of PDAC. 29 January 2019 Method: Transcriptome data mining techniques were used in order to construct the interactome of the Accepted 9 February 2019 AQP gene family and to determine which genes members are differentially expressed in PDAC as Available online 11 February 2019 compared to controls. The same techniques were used in order to evaluate the potential prognostic role of the differentially expressed genes. Keywords: PDAC Results: Transcriptome microarray data of four GEO datasets were incorporated, including 142 primary Aquaporin tumor samples and 104 normal pancreatic tissue samples. -

A Minimum-Labeling Approach for Reconstructing Protein Networks Across Multiple Conditions
A Minimum-Labeling Approach for Reconstructing Protein Networks across Multiple Conditions Arnon Mazza1, Irit Gat-Viks2, Hesso Farhan3, and Roded Sharan1 1 Blavatnik School of Computer Science, Tel Aviv University, Tel Aviv 69978, Israel. Email: [email protected]. 2 Dept. of Cell Research and Immunology, Tel Aviv University, Tel Aviv 69978, Israel. 3 Biotechnology Institute Thurgau, University of Konstanz, Unterseestrasse 47, CH-8280 Kreuzlingen, Switzerland. Abstract. The sheer amounts of biological data that are generated in recent years have driven the development of network analysis tools to fa- cilitate the interpretation and representation of these data. A fundamen- tal challenge in this domain is the reconstruction of a protein-protein sub- network that underlies a process of interest from a genome-wide screen of associated genes. Despite intense work in this area, current algorith- mic approaches are largely limited to analyzing a single screen and are, thus, unable to account for information on condition-specific genes, or reveal the dynamics (over time or condition) of the process in question. Here we propose a novel formulation for network reconstruction from multiple-condition data and devise an efficient integer program solution for it. We apply our algorithm to analyze the response to influenza in- fection in humans over time as well as to analyze a pair of ER export related screens in humans. By comparing to an extant, single-condition tool we demonstrate the power of our new approach in integrating data from multiple conditions in a compact and coherent manner, capturing the dynamics of the underlying processes. 1 Introduction With the increasing availability of high-throughput data, network biol- arXiv:1307.7803v1 [q-bio.QM] 30 Jul 2013 ogy has become the method of choice for filtering, interpreting and rep- resenting these data. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -

Conserved and Novel Properties of Clathrin-Mediated Endocytosis in Dictyostelium Discoideum" (2012)
Rockefeller University Digital Commons @ RU Student Theses and Dissertations 2012 Conserved and Novel Properties of Clathrin- Mediated Endocytosis in Dictyostelium Discoideum Laura Macro Follow this and additional works at: http://digitalcommons.rockefeller.edu/ student_theses_and_dissertations Part of the Life Sciences Commons Recommended Citation Macro, Laura, "Conserved and Novel Properties of Clathrin-Mediated Endocytosis in Dictyostelium Discoideum" (2012). Student Theses and Dissertations. Paper 163. This Thesis is brought to you for free and open access by Digital Commons @ RU. It has been accepted for inclusion in Student Theses and Dissertations by an authorized administrator of Digital Commons @ RU. For more information, please contact [email protected]. CONSERVED AND NOVEL PROPERTIES OF CLATHRIN- MEDIATED ENDOCYTOSIS IN DICTYOSTELIUM DISCOIDEUM A Thesis Presented to the Faculty of The Rockefeller University in Partial Fulfillment of the Requirements for the degree of Doctor of Philosophy by Laura Macro June 2012 © Copyright by Laura Macro 2012 CONSERVED AND NOVEL PROPERTIES OF CLATHRIN- MEDIATED ENDOCYTOSIS IN DICTYOSTELIUM DISCOIDEUM Laura Macro, Ph.D. The Rockefeller University 2012 The protein clathrin mediates one of the major pathways of endocytosis from the extracellular milieu and plasma membrane. Clathrin functions with a network of interacting accessory proteins, one of which is the adaptor complex AP-2, to co-ordinate vesicle formation. Disruption of genes involved in clathrin-mediated endocytosis causes embryonic lethality in multicellular animals suggesting that clathrin-mediated endocytosis is a fundamental cellular process. However, loss of clathrin-mediated endocytosis genes in single cell eukaryotes, such as S.cerevisiae (yeast), does not cause lethality, suggesting that clathrin may convey specific advantages for multicellularity. -
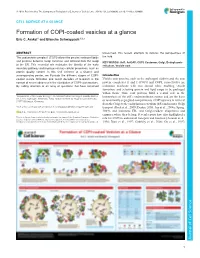
Formation of COPI-Coated Vesicles at a Glance Eric C
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs209890. doi:10.1242/jcs.209890 CELL SCIENCE AT A GLANCE Formation of COPI-coated vesicles at a glance Eric C. Arakel1 and Blanche Schwappach1,2,* ABSTRACT unresolved, this review attempts to refocus the perspectives of The coat protein complex I (COPI) allows the precise sorting of lipids the field. and proteins between Golgi cisternae and retrieval from the Golgi KEY WORDS: Arf1, ArfGAP, COPI, Coatomer, Golgi, Endoplasmic to the ER. This essential role maintains the identity of the early reticulum, Vesicle coat secretory pathway and impinges on key cellular processes, such as protein quality control. In this Cell Science at a Glance and accompanying poster, we illustrate the different stages of COPI- Introduction coated vesicle formation and revisit decades of research in the Vesicle coat proteins, such as the archetypal clathrin and the coat context of recent advances in the elucidation of COPI coat structure. protein complexes II and I (COPII and COPI, respectively) are By calling attention to an array of questions that have remained molecular machines with two central roles: enabling vesicle formation, and selecting protein and lipid cargo to be packaged within them. Thus, coat proteins fulfil a central role in the 1Department of Molecular Biology, Universitätsmedizin Göttingen, Humboldtallee homeostasis of the cell’s endomembrane system and are the basis 23, 37073 Göttingen, Germany. 2Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. of functionally segregated compartments. COPI operates in retrieval from the Golgi to the endoplasmic reticulum (ER) and in intra-Golgi *Author for correspondence ([email protected]) transport (Beck et al., 2009; Duden, 2003; Lee et al., 2004a; Spang, E.C.A., 0000-0001-7716-7149; B.S., 0000-0003-0225-6432 2009), and maintains ER- and Golgi-resident chaperones and enzymes where they belong. -

Supplementary Figures 1-14 and Supplementary References
SUPPORTING INFORMATION Spatial Cross-Talk Between Oxidative Stress and DNA Replication in Human Fibroblasts Marko Radulovic,1,2 Noor O Baqader,1 Kai Stoeber,3† and Jasminka Godovac-Zimmermann1* 1Division of Medicine, University College London, Center for Nephrology, Royal Free Campus, Rowland Hill Street, London, NW3 2PF, UK. 2Insitute of Oncology and Radiology, Pasterova 14, 11000 Belgrade, Serbia 3Research Department of Pathology and UCL Cancer Institute, Rockefeller Building, University College London, University Street, London WC1E 6JJ, UK †Present Address: Shionogi Europe, 33 Kingsway, Holborn, London WC2B 6UF, UK TABLE OF CONTENTS 1. Supplementary Figures 1-14 and Supplementary References. Figure S-1. Network and joint spatial razor plot for 18 enzymes of glycolysis and the pentose phosphate shunt. Figure S-2. Correlation of SILAC ratios between OXS and OAC for proteins assigned to the SAME class. Figure S-3. Overlap matrix (r = 1) for groups of CORUM complexes containing 19 proteins of the 49-set. Figure S-4. Joint spatial razor plots for the Nop56p complex and FIB-associated complex involved in ribosome biogenesis. Figure S-5. Analysis of the response of emerin nuclear envelope complexes to OXS and OAC. Figure S-6. Joint spatial razor plots for the CCT protein folding complex, ATP synthase and V-Type ATPase. Figure S-7. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated by GO to nucleocytoplasmic transport (GO:0006913). Figure S-8. Joint spatial razor plots showing changes in subcellular abundance and compartmental distribution for proteins annotated to endocytosis (GO:0006897). Figure S-9. Joint spatial razor plots for 401-set proteins annotated by GO to small GTPase mediated signal transduction (GO:0007264) and/or GTPase activity (GO:0003924). -

Overlapping Role of SCYL1 and SCYL3 in Maintaining Motor Neuron Viability
The Journal of Neuroscience, March 7, 2018 • 38(10):2615–2630 • 2615 Neurobiology of Disease Overlapping Role of SCYL1 and SCYL3 in Maintaining Motor Neuron Viability Emin Kuliyev,1 Sebastien Gingras,4 XClifford S. Guy,1 Sherie Howell,2 Peter Vogel,3 and XStephane Pelletier1 1Departments of Immunology, 2Pathology, 3Veterinary Pathology Core, Advanced Histology Core, St. Jude Children’s Research Hospital, Memphis, Tennessee 38105, and 4Department of Immunology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania 15213 Members of the SCY1-like (SCYL) family of protein kinases are evolutionarily conserved and ubiquitously expressed proteins character- ized by an N-terminal pseudokinase domain, centrally located Huntingtin, elongation factor 3, protein phosphatase 2A, yeast kinase TOR1 repeats, and an overall disorganized C-terminal segment. In mammals, three family members encoded by genes Scyl1, Scyl2, and Scyl3 have been described. Studies have pointed to a role for SCYL1 and SCYL2 in regulating neuronal function and viability in mice and humans, but little is known about the biological function of SCYL3. Here, we show that the biochemical and cell biological properties of SCYL3aresimilartothoseofSCYL1andbothproteinsworkinconjunctiontomaintainmotorneuronviability.Specifically,althoughlack of Scyl3 in mice has no apparent effect on embryogenesis and postnatal life, it accelerates the onset of the motor neuron disorder caused by Scyl1 deficiency. Growth abnormalities, motor dysfunction, hindlimb paralysis, muscle wasting, neurogenic atrophy, motor neuron degeneration, and loss of large-caliber axons in peripheral nerves occurred at an earlier age in Scyl1/Scyl3 double-deficient mice than in Scyl1-deficientmice.DiseaseonsetalsocorrelatedwiththemislocalizationofTDP-43inspinalmotorneurons,suggestingthatSCYL1and SCYL3 regulate TDP-43 proteostasis. Together, our results demonstrate an overlapping role for SCYL1 and SCYL3 in vivo and highlight the importance the SCYL family of proteins in regulating neuronal function and survival. -

Anti-COPE Picoband Antibody Catalog # ABO13033
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Anti-COPE Picoband Antibody Catalog # ABO13033 Specification Anti-COPE Picoband Antibody - Product Information Application IHC Primary Accession O14579 Host Rabbit Reactivity Human, Mouse, Rat Clonality Polyclonal Format Lyophilized Description Rabbit IgG polyclonal antibody for Coatomer subunit epsilon(COPE) detection. Tested with WB, IHC-P in Human;Mouse;Rat. Figure 4. IHC analysis of COPE using Reconstitution anti-COPE antibody (ABO13033). Add 0.2ml of distilled water will yield a concentration of 500ug/ml. Anti-COPE Picoband Antibody - Additional Information Gene ID 11316 Other Names Coatomer subunit epsilon, Epsilon-coat protein, Epsilon-COP, COPE Calculated MW 34482 MW KDa Application Details Immunohistochemistry(Paraffin-embedded Section), 0.5-1 µg/ml, Human, Mouse, Rat, By Heat<br> Western blot, 0.1-0.5 µg/ml, Human, Mouse, Rat, <br> <br> Subcellular Localization Cytoplasm . Golgi apparatus membrane ; Peripheral membrane protein ; Cytoplasmic side . Cytoplasmic vesicle, COPI-coated vesicle membrane ; Peripheral membrane protein ; Cytoplasmic side . The coatomer is cytoplasmic or polymerized on the cytoplasmic side of the Golgi, as well as on the vesicles/buds originating from it. Page 1/3 10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Contents Each vial contains 5mg BSA, 0.9mg NaCl, 0.2mg Na2HPO4, 0.05mg NaN3. Immunogen E. coli-derived human COPE recombinant protein (Position: E80-A308). Human COPE shares 89.5% amino acid (aa) sequence identity with mouse COPE. Purification Immunogen affinity purified. Cross Reactivity No cross reactivity with other proteins.