Complete Genome Sequence of the Polysaccharide-Degrading Rumen
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Effects of Dietary Supplementation with Hainanmycin on Protein Degradation and Populations of Ammonia-Producing Bacteria in Vitro
668 Asian Australas. J. Anim. Sci. Vol. 26, No. 5 : 668-674 May 2013 http://dx.doi.org/10.5713/ajas.2012.12589 www.ajas.info pISSN 1011-2367 eISSN 1976-5517 Effects of Dietary Supplementation with Hainanmycin on Protein Degradation and Populations of Ammonia-producing Bacteria In vitro Z. B. Wang1,2,a, H. S. Xin1,a, M. J. Wang1, Z. Y. Li1, Y. L. Qu2, S. J. Miao2 and Y. G. Zhang1,* 1 College of Animal Science and Technology, Northeast Agricultural University, Harbin, 150030, Heilongjiang, China ABSTRACT: An in vitro fermentation was conducted to determine the effects of hainanmycin on protein degradation and populations of ammonia-producing bacteria. The substrates (DM basis) for in vitro fermentation consisted of alfalfa hay (31.7%), Chinese wild rye grass hay (28.3%), ground corn grain (24.5%), soybean meal (15.5%) with a forage: concentrate of 60:40. Treatments were the control (no additive) and hainanmycin supplemented at 0.1 (H0.1), 1 (H1), 10 (H10), and 100 mg/kg (H100) of the substrates. After 24 h of fermentation, the highest addition level of hainanmycin decreased total VFA concentration and increased the final pH. The high addition level of hainanmycin (H1, H10, and H100) reduced (p<0.05) branched-chain VFA concentration, the molar proportion of acetate and butyrate, and ratio of acetate to propionate; and increased the molar proportion of propionate, except that for H1 the in molar proportion of acetate and isobutyrate was not changed (p>0.05). After 24 h of fermentation, H10 and H100 increased (p<0.05) concentrations of peptide nitrogen and AA nitrogen and proteinase activity, and decreased (p<0.05) NH3-N concentration and deaminase activity compared with control. -

Comparison of Enzymatic Activities and Proteomic Profiles Of
Sechovcová et al. Proteome Science (2019) 17:2 https://doi.org/10.1186/s12953-019-0150-3 RESEARCH Open Access Comparison of enzymatic activities and proteomic profiles of Butyrivibrio fibrisolvens grown on different carbon sources Hana Sechovcová1,5* , Lucie Kulhavá2,4, Kateřina Fliegerová1, Mária Trundová3, Daniel Morais6, Jakub Mrázek1 and Jan Kopečný1 Abstract Background: The rumen microbiota is one of the most complex consortia of anaerobes, involving archaea, bacteria, protozoa, fungi and phages. They are very effective at utilizing plant polysaccharides, especially cellulose and hemicelluloses. The most important hemicellulose decomposers are clustered with the genus Butyrivibrio.As the related species differ in their range of hydrolytic activities and substrate preferences, Butyrivibrio fibrisolvens was selected as one of the most effective isolates and thus suitable for proteomic studies on substrate comparisons in the extracellular fraction. The B. fibrisolvens genome is the biggest in the butyrivibria cluster and is focused on “environmental information processing” and “carbohydrate metabolism”. Methods: The study of the effect of carbon source on B. fibrisolvens 3071 was based on cultures grown on four substrates: xylose, glucose, xylan, xylan with 25% glucose. The enzymatic activities were studied by spectrophotometric and zymogram methods. Proteomic study was based on genomics, 2D electrophoresis and nLC/MS (Bruker Daltonics) analysis. Results: Extracellular β-endoxylanase as well as xylan β-xylosidase activities were induced with xylan. The presence of the xylan polymer induced hemicellulolytic enzymes and increased the protein fraction in the interval from 40 to 80 kDa. 2D electrophoresis with nLC/MS analysis of extracellular B. fibrisolvens 3071 proteins found 14 diverse proteins with significantly different expression on the tested substrates. -

Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis
International Journal of Molecular Sciences Review Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis David Johane Machate 1, Priscila Silva Figueiredo 2 , Gabriela Marcelino 2 , Rita de Cássia Avellaneda Guimarães 2,*, Priscila Aiko Hiane 2 , Danielle Bogo 2, Verônica Assalin Zorgetto Pinheiro 2, Lincoln Carlos Silva de Oliveira 3 and Arnildo Pott 1 1 Graduate Program in Biotechnology and Biodiversity in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] (D.J.M.); [email protected] (A.P.) 2 Graduate Program in Health and Development in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; pri.fi[email protected] (P.S.F.); [email protected] (G.M.); [email protected] (P.A.H.); [email protected] (D.B.); [email protected] (V.A.Z.P.) 3 Chemistry Institute, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-67-3345-7416 Received: 9 March 2020; Accepted: 27 March 2020; Published: 8 June 2020 Abstract: Long-term high-fat dietary intake plays a crucial role in the composition of gut microbiota in animal models and human subjects, which affect directly short-chain fatty acid (SCFA) production and host health. This review aims to highlight the interplay of fatty acid (FA) intake and gut microbiota composition and its interaction with hosts in health promotion and obesity prevention and its related metabolic dysbiosis. -

Biotechnological Approaches to Manure Nutrient Management
COUNCIL FORFOR AGRICULTURALAGRICULTURAL SCIENCE SCIENCE AND AND TECHNOLOGY—1 TECHNOLOGY NUMBER 33 JULY 2006 ANIMAL AGRICULTURE'S FUTURE THROUGH BIOTECHNOLOGY, PART 4 BIOTECHNOLOGICAL APPROACHES TO MANURE NUTRIENT MANAGEMENT TASK FORCE MEMBERS: Xingen Lei, Chair, Department of Animal Science, Cornell Univer- neering. This type of technol- INTRODUCTION sity, Ithaca, New York; John P. Blake, Depart- Food animals are fed and ogy may be applied to de- produced for the purpose of ment of Poultry Science, Auburn University, crease total manure mass or feeding humans. Manure from Auburn, Alabama; Cecil W. Forsberg, Depart- concentrations of P, N, am- these animals is a valuable ment of Microbiology, University of Guelph, monia (NH3), trace elements, source of fertilizer, but con- Ontario, Canada; Danny G. Fox, Department of and other factors. Targeted modifications centrations of manure nutri- Animal Science, Cornell University, Ithaca, New 1 can be based strategically on ents such as phosphorus (P) , York; Elizabeth Grabau, Department of Plant nitrogen (N), and metals may plants, animals, ruminal and Pathology, Physiology, and Weed Science, Vir- exceed needs for plant growth intestinal microorganisms, and cause environmental pol- ginia Tech, Blacksburg; Zdzislaw Mroz, Ani- and diets. The plant-based lution. Thus, managing live- mal Sciences Group, Division of Nutrition and approaches include genetic stock manure nutrients has Food, Wageningen, The Netherlands; Alan L. and chemical modifications of been a task shared by those Sutton, Department of Animal Sciences, Purdue feeds (e.g., overexpressing hydrolytic enzymes such as interested in animal nutrition, University, West Lafayette, Indiana; William R. agronomy, and environmental phytase in seeds, decreasing Walker, DPI Global, Fort Dodge, Iowa; Ken- protection. -

The Isolation of Novel Lachnospiraceae Strains and the Evaluation of Their Potential Roles in Colonization Resistance Against Clostridium Difficile
The isolation of novel Lachnospiraceae strains and the evaluation of their potential roles in colonization resistance against Clostridium difficile Diane Yuan Wang Honors Thesis in Biology Department of Ecology and Evolutionary Biology College of Literature, Science, & the Arts University of Michigan, Ann Arbor April 1st, 2014 Sponsor: Vincent B. Young, M.D., Ph.D. Associate Professor of Internal Medicine Associate Professor of Microbiology and Immunology Medical School Co-Sponsor: Aaron A. King, Ph.D. Associate Professor of Ecology & Evolutionary Associate Professor of Mathematics College of Literature, Science, & the Arts Reader: Blaise R. Boles, Ph.D. Assistant Professor of Molecular, Cellular and Developmental Biology College of Literature, Science, & the Arts 1 Table of Contents Abstract 3 Introduction 4 Clostridium difficile 4 Colonization Resistance 5 Lachnospiraceae 6 Objectives 7 Materials & Methods 9 Sample Collection 9 Bacterial Isolation and Selective Growth Conditions 9 Design of Lachnospiraceae 16S rRNA-encoding gene primers 9 DNA extraction and 16S ribosomal rRNA-encoding gene sequencing 10 Phylogenetic analyses 11 Direct inhibition 11 Bile salt hydrolase (BSH) detection 12 PCR assay for bile acid 7α-dehydroxylase detection 12 Tables & Figures Table 1 13 Table 2 15 Table 3 21 Table 4 25 Figure 1 16 Figure 2 19 Figure 3 20 Figure 4 24 Figure 5 26 Results 14 Isolation of novel Lachnospiraceae strains 14 Direct inhibition 17 Bile acid physiology 22 Discussion 27 Acknowledgments 33 References 34 2 Abstract Background: Antibiotic disruption of the gastrointestinal tract’s indigenous microbiota can lead to one of the most common nosocomial infections, Clostridium difficile, which has an annual cost exceeding $4.8 billion dollars. -

Breast Milk Microbiota: a Review of the Factors That Influence Composition
Published in "Journal of Infection 81(1): 17–47, 2020" which should be cited to refer to this work. ✩ Breast milk microbiota: A review of the factors that influence composition ∗ Petra Zimmermann a,b,c,d, , Nigel Curtis b,c,d a Department of Paediatrics, Fribourg Hospital HFR and Faculty of Science and Medicine, University of Fribourg, Switzerland b Department of Paediatrics, The University of Melbourne, Parkville, Australia c Infectious Diseases Research Group, Murdoch Children’s Research Institute, Parkville, Australia d Infectious Diseases Unit, The Royal Children’s Hospital Melbourne, Parkville, Australia s u m m a r y Breastfeeding is associated with considerable health benefits for infants. Aside from essential nutrients, immune cells and bioactive components, breast milk also contains a diverse range of microbes, which are important for maintaining mammary and infant health. In this review, we summarise studies that have Keywords: investigated the composition of the breast milk microbiota and factors that might influence it. Microbiome We identified 44 studies investigating 3105 breast milk samples from 2655 women. Several studies Diversity reported that the bacterial diversity is higher in breast milk than infant or maternal faeces. The maxi- Delivery mum number of each bacterial taxonomic level detected per study was 58 phyla, 133 classes, 263 orders, Caesarean 596 families, 590 genera, 1300 species and 3563 operational taxonomic units. Furthermore, fungal, ar- GBS chaeal, eukaryotic and viral DNA was also detected. The most frequently found genera were Staphylococ- Antibiotics cus, Streptococcus Lactobacillus, Pseudomonas, Bifidobacterium, Corynebacterium, Enterococcus, Acinetobacter, BMI Rothia, Cutibacterium, Veillonella and Bacteroides. There was some evidence that gestational age, delivery Probiotics mode, biological sex, parity, intrapartum antibiotics, lactation stage, diet, BMI, composition of breast milk, Smoking Diet HIV infection, geographic location and collection/feeding method influence the composition of the breast milk microbiota. -

Metagenomic Surveys of Gut Microbiota
Accepted Manuscript Metagenomic Surveys of Gut Microbiota Rahul Shubhra Mandal, Sudipto Saha, Santasabuj Das PII: S1672-0229(15)00054-6 DOI: http://dx.doi.org/10.1016/j.gpb.2015.02.005 Reference: GPB 159 To appear in: Genomics, Proteomics & Bioinformatics Received Date: 8 July 2014 Revised Date: 10 February 2015 Accepted Date: 26 February 2015 Please cite this article as: R.S. Mandal, S. Saha, S. Das, Metagenomic Surveys of Gut Microbiota, Genomics, Proteomics & Bioinformatics (2015), doi: http://dx.doi.org/10.1016/j.gpb.2015.02.005 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. 1 Metagenomic Surveys of Gut Microbiota 2 3 Rahul Shubhra Mandal1,a, Sudipto Saha2,*,b, Santasabuj Das1,3,*,c 4 5 1Biomedical Informatics Centre, National Institute of Cholera and Enteric Diseases, Kolkata 6 700010, India 7 2Bioinformatics Centre, Bose Institute, Kolkata 700054, India 8 3Division of Clinical Medicine, National Institute of Cholera and Enteric Diseases, Kolkata 9 700010, India 10 11 *Corresponding authors. 12 E-mail: [email protected] (Das S), [email protected] (Saha S). 13 Running title: Mandal RS et al / Metagenomic Analysis of Gut Microbiota 14 15 aORCID: 0000-0001-8939-3362. -

Characterization of Antibiotic Resistance Genes in the Species of the Rumen Microbiota
ARTICLE https://doi.org/10.1038/s41467-019-13118-0 OPEN Characterization of antibiotic resistance genes in the species of the rumen microbiota Yasmin Neves Vieira Sabino1, Mateus Ferreira Santana1, Linda Boniface Oyama2, Fernanda Godoy Santos2, Ana Júlia Silva Moreira1, Sharon Ann Huws2* & Hilário Cuquetto Mantovani 1* Infections caused by multidrug resistant bacteria represent a therapeutic challenge both in clinical settings and in livestock production, but the prevalence of antibiotic resistance genes 1234567890():,; among the species of bacteria that colonize the gastrointestinal tract of ruminants is not well characterized. Here, we investigate the resistome of 435 ruminal microbial genomes in silico and confirm representative phenotypes in vitro. We find a high abundance of genes encoding tetracycline resistance and evidence that the tet(W) gene is under positive selective pres- sure. Our findings reveal that tet(W) is located in a novel integrative and conjugative element in several ruminal bacterial genomes. Analyses of rumen microbial metatranscriptomes confirm the expression of the most abundant antibiotic resistance genes. Our data provide insight into antibiotic resistange gene profiles of the main species of ruminal bacteria and reveal the potential role of mobile genetic elements in shaping the resistome of the rumen microbiome, with implications for human and animal health. 1 Departamento de Microbiologia, Universidade Federal de Viçosa, Viçosa, Minas Gerais, Brazil. 2 Institute for Global Food Security, School of Biological -

Electron Transport Phosphorylation in Rumen Butyrivibrios: Unprecedented ATP Yield for Glucose Fermentation to Butyrate
HYPOTHESIS AND THEORY published: 24 June 2015 doi: 10.3389/fmicb.2015.00622 Electron transport phosphorylation in rumen butyrivibrios: unprecedented ATP yield for glucose fermentation to butyrate Timothy J. Hackmann1 and Jeffrey L. Firkins2* 1 Department of Animal Sciences, University of Florida, Gainesville, FL, USA, 2 Department of Animal Sciences, The Ohio State University, Columbus, OH, USA From a genomic analysis of rumen butyrivibrios (Butyrivibrio and Pseudobutyrivibrio sp.), we have re-evaluated the contribution of electron transport phosphorylation (ETP) to ATP formation in this group. This group is unique in that most (76%) genomes were predicted to possess genes for both Ech and Rnf transmembrane ion pumps. These pumps act in concert with the NifJ and Bcd-Etf to form a electrochemical potential (μH+ and μNa+), which drives ATP synthesis by ETP. Of the 62 total butyrivibrio genomes currently available from the Hungate 1000 project, all 62 were predicted to Edited by: possess NifJ, which reduces oxidized ferredoxin (Fdox) during pyruvate conversion to Emilio M. Ungerfeld, Instituto de Investigaciones acetyl-CoA. All 62 possessed all subunits of Bcd-Etf, which reduces Fdox and oxidizes Agropecuarias, Chile reduced NAD during crotonyl-CoA reduction. Additionally, 61 genomes possessed all Reviewed by: subunits of the Rnf, which generates μH+ or μNa+ from oxidation of reduced Fd Wolfgang Buckel, (Fdred) and reduction of oxidized NAD. Further, 47 genomes possessed all six subunits Philipps-Universität Marburg, + Germany of the Ech, which generates μH from oxidation of Fdred. For glucose fermentation Robert J. Wallace, to butyrate and H2, the electrochemical potential established should drive synthesis University of Aberdeen, UK of ∼1.5 ATP by the F0F1-ATP synthase (possessed by all 62 genomes). -

Supplementary Informations SI2. Supplementary Table 1
Supplementary Informations SI2. Supplementary Table 1. M9, soil, and rhizosphere media composition. LB in Compound Name Exchange Reaction LB in soil LBin M9 rhizosphere H2O EX_cpd00001_e0 -15 -15 -10 O2 EX_cpd00007_e0 -15 -15 -10 Phosphate EX_cpd00009_e0 -15 -15 -10 CO2 EX_cpd00011_e0 -15 -15 0 Ammonia EX_cpd00013_e0 -7.5 -7.5 -10 L-glutamate EX_cpd00023_e0 0 -0.0283302 0 D-glucose EX_cpd00027_e0 -0.61972444 -0.04098397 0 Mn2 EX_cpd00030_e0 -15 -15 -10 Glycine EX_cpd00033_e0 -0.0068175 -0.00693094 0 Zn2 EX_cpd00034_e0 -15 -15 -10 L-alanine EX_cpd00035_e0 -0.02780553 -0.00823049 0 Succinate EX_cpd00036_e0 -0.0056245 -0.12240603 0 L-lysine EX_cpd00039_e0 0 -10 0 L-aspartate EX_cpd00041_e0 0 -0.03205557 0 Sulfate EX_cpd00048_e0 -15 -15 -10 L-arginine EX_cpd00051_e0 -0.0068175 -0.00948672 0 L-serine EX_cpd00054_e0 0 -0.01004986 0 Cu2+ EX_cpd00058_e0 -15 -15 -10 Ca2+ EX_cpd00063_e0 -15 -100 -10 L-ornithine EX_cpd00064_e0 -0.0068175 -0.00831712 0 H+ EX_cpd00067_e0 -15 -15 -10 L-tyrosine EX_cpd00069_e0 -0.0068175 -0.00233919 0 Sucrose EX_cpd00076_e0 0 -0.02049199 0 L-cysteine EX_cpd00084_e0 -0.0068175 0 0 Cl- EX_cpd00099_e0 -15 -15 -10 Glycerol EX_cpd00100_e0 0 0 -10 Biotin EX_cpd00104_e0 -15 -15 0 D-ribose EX_cpd00105_e0 -0.01862144 0 0 L-leucine EX_cpd00107_e0 -0.03596182 -0.00303228 0 D-galactose EX_cpd00108_e0 -0.25290619 -0.18317325 0 L-histidine EX_cpd00119_e0 -0.0068175 -0.00506825 0 L-proline EX_cpd00129_e0 -0.01102953 0 0 L-malate EX_cpd00130_e0 -0.03649016 -0.79413596 0 D-mannose EX_cpd00138_e0 -0.2540567 -0.05436649 0 Co2 EX_cpd00149_e0 -
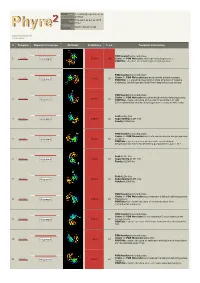
Phyre 2 Results for P25553
Email [email protected] Description P25553 Thu Jan 5 11:42:11 GMT Date 2012 Unique Job 5024f4b9e5342484 ID Detailed template information # Template Alignment Coverage 3D Model Confidence % i.d. Template Information PDB header:oxidoreductase 1 c2hg2A_ Alignment 100.0 100 Chain: A: PDB Molecule:aldehyde dehydrogenase a; PDBTitle: structure of lactaldehyde dehydrogenase PDB header:oxidoreductase Chain: B: PDB Molecule:betaine aldehyde dehydrogenase; 2 c3ed6B_ 100.0 36 Alignment PDBTitle: 1.7 angstrom resolution crystal structure of betaine aldehyde2 dehydrogenase (betb) from staphylococcus aureus PDB header:oxidoreductase Chain: A: PDB Molecule:formyltetrahydrofolate dehydrogenase; 3 c2o2qA_ 100.0 33 Alignment PDBTitle: crystal structure of the c-terminal domain of rat2 10'formyltetrahydrofolate dehydrogenase in complex with nadp Fold:ALDH-like 4 d1a4sa_ Alignment 100.0 35 Superfamily:ALDH-like Family:ALDH-like PDB header:oxidoreductase Chain: H: PDB Molecule:succinate-semialdehyde dehydrogenase 5 c3ifgH_ Alignment 100.0 34 (nadp+); PDBTitle: crystal structure of succinate-semialdehyde dehydrogenase from2 burkholderia pseudomallei, part 1 of 2 Fold:ALDH-like 6 d1bxsa_ Alignment 100.0 33 Superfamily:ALDH-like Family:ALDH-like Fold:ALDH-like 7 d1o9ja_ Alignment 100.0 33 Superfamily:ALDH-like Family:ALDH-like PDB header:oxidoreductase Chain: A: PDB Molecule:succinate-semialdehyde dehydrogenase 8 c3rh9A_ Alignment 100.0 35 (nad(p)(+)); PDBTitle: the crystal structure of oxidoreductase from marinobacter aquaeolei PDB header:oxidoreductase Chain: B: PDB Molecule:5-carboxymethyl-2-hydroxymuconate 9 c2d4eB_ Alignment 100.0 32 semialdehyde PDBTitle: crystal structure of the hpcc from thermus thermophilus hb8 PDB header:oxidoreductase Chain: G: PDB Molecule:antiquitin; 10 c2jg7G_ 100.0 28 Alignment PDBTitle: crystal structure of seabream antiquitin and elucidation of2 its substrate specificity PDB header:oxidoreductase Chain: C: PDB Molecule:succinate-semialdehyde dehydrogenase 11 c3jz4C_ 100.0 39 Alignment [nadp+]; PDBTitle: crystal structure of e. -

Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome
Research JAMA Pediatrics | Original Investigation Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome Pia S. Pannaraj, MD, MPH; Fan Li, PhD; Chiara Cerini, MD; Jeffrey M. Bender, MD; Shangxin Yang, PhD; Adrienne Rollie, MS; Helty Adisetiyo, PhD; Sara Zabih, MS; Pamela J. Lincez, PhD; Kyle Bittinger, PhD; Aubrey Bailey, MS; Frederic D. Bushman, PhD; John W. Sleasman, MD; Grace M. Aldrovandi, MD Supplemental content IMPORTANCE Establishment of the infant microbiome has lifelong implications on health and immunity. Gut microbiota of breastfed compared with nonbreastfed individuals differ during infancy as well as into adulthood. Breast milk contains a diverse population of bacteria, but little is known about the vertical transfer of bacteria from mother to infant by breastfeeding. OBJECTIVE To determine the association between the maternal breast milk and areolar skin and infant gut bacterial communities. DESIGN, SETTING, AND PARTICIPANTS In a prospective, longitudinal study, bacterial composition was identified with sequencing of the 16S ribosomal RNA gene in breast milk, areolar skin, and infant stool samples of 107 healthy mother-infant pairs. The study was conducted in Los Angeles, California, and St Petersburg, Florida, between January 1, 2010, and February 28, 2015. EXPOSURES Amount and duration of daily breastfeeding and timing of solid food introduction. MAIN OUTCOMES AND MEASURES Bacterial composition in maternal breast milk, areolar skin, and infant stool by sequencing of the 16S ribosomal RNA gene. RESULTS In the 107 healthy mother and infant pairs (median age at the time of specimen collection, 40 days; range, 1-331 days), 52 (43.0%) of the infants were male.