Comparative Genomic Analysis of Integral Membrane Transport Proteins in Ciliates
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Single-Particle Cryo-EM of the Ryanodine Receptor Channel in an Aqueous Environment
Single-particle cryo-EM of the ryanodine receptor channel Eur J Transl Myol - Basic Appl Myol 2015; 25 (1): 35-48 Single-particle cryo-EM of the ryanodine receptor channel in an aqueous environment Mariah R. Baker, Guizhen Fan and Irina I. Serysheva Department of Biochemistry and Molecular Biology, The University of Texas Medical School at Houston, 6431 Fannin Street, Houston, TX 77030, USA Abstract Ryanodine receptors (RyRs) are tetrameric ligand-gated Ca2+ release channels that are responsible for the increase of cytosolic Ca2+ concentration leading to muscle contraction. Our current understanding of RyR channel gating and regulation is greatly limited due to the lack of a high-resolution structure of the channel protein. The enormous size and unwieldy shape of Ca2+ release channels make X-ray or NMR methods difficult to apply for high-resolution structural analysis of the full-length functional channel. Single-particle electron cryo- microscopy (cryo-EM) is one of the only effective techniques for the study of such a large integral membrane protein and its molecular interactions. Despite recent developments in cryo- EM technologies and break-through single-particle cryo-EM studies of ion channels, cryospecimen preparation, particularly the presence of detergent in the buffer, remains the main impediment to obtaining atomic-resolution structures of ion channels and a multitude of other integral membrane protein complexes. In this review we will discuss properties of several detergents that have been successfully utilized in cryo-EM studies of ion channels and the emergence of the detergent alternative amphipol to stabilize ion channels for structure- function characterization. Future structural studies of challenging specimen like ion channels are likely to be facilitated by cryo-EM amenable detergents or alternative surfactants. -

Minding the Calcium Store: Ryanodine Receptor Activation As a Convergent Mechanism of PCB Toxicity
Pharmacology & Therapeutics 125 (2010) 260–285 Contents lists available at ScienceDirect Pharmacology & Therapeutics journal homepage: www.elsevier.com/locate/pharmthera Associate Editor: Carey Pope Minding the calcium store: Ryanodine receptor activation as a convergent mechanism of PCB toxicity Isaac N. Pessah ⁎, Gennady Cherednichenko, Pamela J. Lein Department of Molecular Biosciences, School of Veterinary Medicine, University of California, Davis, CA 95616, USA article info abstract Keywords: Chronic low-level polychlorinated biphenyl (PCB) exposures remain a significant public health concern since Ryanodine receptor (RyR) results from epidemiological studies indicate that PCB burden is associated with immune system Calcium-induced calcium release dysfunction, cardiovascular disease, and impairment of the developing nervous system. Of these various Calcium regulation adverse health effects, developmental neurotoxicity has emerged as a particularly vulnerable endpoint in Polychlorinated biphenyls PCB toxicity. Arguably the most pervasive biological effects of PCBs could be mediated by their ability to alter Triclosan fi 2+ Bastadins the spatial and temporal delity of Ca signals through one or more receptor-mediated processes. This Polybrominated diphenylethers review will focus on our current knowledge of the structure and function of ryanodine receptors (RyRs) in Developmental neurotoxicity muscle and nerve cells and how PCBs and related non-coplanar structures alter these functions. The Activity dependent plasticity molecular and cellular mechanisms by which non-coplanar PCBs and related structures alter local and global Ca2+ signaling properties and the possible short and long-term consequences of these perturbations on neurodevelopment and neurodegeneration are reviewed. © 2009 Elsevier Inc. All rights reserved. Contents 1. Introduction ............................................... 260 2. Ryanodine receptor macromolecular complexes: significance to polychlorinated biphenyl-mediated Ca2+ dysregulation . -

Snapshot: Mammalian TRP Channels David E
SnapShot: Mammalian TRP Channels David E. Clapham HHMI, Children’s Hospital, Department of Neurobiology, Harvard Medical School, Boston, MA 02115, USA TRP Activators Inhibitors Putative Interacting Proteins Proposed Functions Activation potentiated by PLC pathways Gd, La TRPC4, TRPC5, calmodulin, TRPC3, Homodimer is a purported stretch-sensitive ion channel; form C1 TRPP1, IP3Rs, caveolin-1, PMCA heteromeric ion channels with TRPC4 or TRPC5 in neurons -/- Pheromone receptor mechanism? Calmodulin, IP3R3, Enkurin, TRPC6 TRPC2 mice respond abnormally to urine-based olfactory C2 cues; pheromone sensing 2+ Diacylglycerol, [Ca ]I, activation potentiated BTP2, flufenamate, Gd, La TRPC1, calmodulin, PLCβ, PLCγ, IP3R, Potential role in vasoregulation and airway regulation C3 by PLC pathways RyR, SERCA, caveolin-1, αSNAP, NCX1 La (100 µM), calmidazolium, activation [Ca2+] , 2-APB, niflumic acid, TRPC1, TRPC5, calmodulin, PLCβ, TRPC4-/- mice have abnormalities in endothelial-based vessel C4 i potentiated by PLC pathways DIDS, La (mM) NHERF1, IP3R permeability La (100 µM), activation potentiated by PLC 2-APB, flufenamate, La (mM) TRPC1, TRPC4, calmodulin, PLCβ, No phenotype yet reported in TRPC5-/- mice; potentially C5 pathways, nitric oxide NHERF1/2, ZO-1, IP3R regulates growth cones and neurite extension 2+ Diacylglycerol, [Ca ]I, 20-HETE, activation 2-APB, amiloride, Cd, La, Gd Calmodulin, TRPC3, TRPC7, FKBP12 Missense mutation in human focal segmental glomerulo- C6 potentiated by PLC pathways sclerosis (FSGS); abnormal vasoregulation in TRPC6-/- -

Ca Signaling in Cardiac Fibroblasts and Fibrosis-Associated Heart
Journal of Cardiovascular Development and Disease Review Ca2+ Signaling in Cardiac Fibroblasts and Fibrosis-Associated Heart Diseases Jianlin Feng 1, Maria K. Armillei 1, Albert S. Yu 1, Bruce T. Liang 1, Loren W. Runnels 2,* and Lixia Yue 1,* 1 Calhoun Cardiology Center, Department of Cell Biology, University of Connecticut Health Center, Farmington, CT 06030, USA; [email protected] (J.F.); [email protected] (M.K.A.); [email protected] (A.S.Y.); [email protected] (B.T.L.) 2 Department of Pharmacology, Rutgers, Robert Wood Johnson Medical School, Piscataway, NJ 08854, USA * Correspondence: [email protected] (L.W.R.); [email protected] (L.Y.) Received: 11 August 2019; Accepted: 18 September 2019; Published: 23 September 2019 Abstract: Cardiac fibrosis is the excessive deposition of extracellular matrix proteins by cardiac fibroblasts and myofibroblasts, and is a hallmark feature of most heart diseases, including arrhythmia, hypertrophy, and heart failure. This maladaptive process occurs in response to a variety of stimuli, including myocardial injury, inflammation, and mechanical overload. There are multiple signaling pathways and various cell types that influence the fibrogenesis cascade. Fibroblasts and myofibroblasts are central effectors. Although it is clear that Ca2+ signaling plays a vital role in this pathological process, what contributes to Ca2+ signaling in fibroblasts and myofibroblasts is still not wholly understood, chiefly because of the large and diverse number of receptors, transporters, and ion channels that influence intracellular Ca2+ signaling. Intracellular Ca2+ signals are generated by Ca2+ release from intracellular Ca2+ stores and by Ca2+ entry through a multitude of Ca2+-permeable ion channels in the plasma membrane. -

Preclinical Model Systems of Ryanodine Receptor 1-Related Myopathies and Malignant Hyperthermia
Lawal et al. Orphanet Journal of Rare Diseases (2020) 15:113 https://doi.org/10.1186/s13023-020-01384-x REVIEW Open Access Preclinical model systems of ryanodine receptor 1-related myopathies and malignant hyperthermia: a comprehensive scoping review of works published 1990– 2019 Tokunbor A. Lawal1, Emily S. Wires2, Nancy L. Terry3, James J. Dowling4 and Joshua J. Todd1* Abstract Background: Pathogenic variations in the gene encoding the skeletal muscle ryanodine receptor (RyR1) are associated with malignant hyperthermia (MH) susceptibility, a life-threatening hypermetabolic condition and RYR1- related myopathies (RYR1-RM), a spectrum of rare neuromuscular disorders. In RYR1-RM, intracellular calcium dysregulation, post-translational modifications, and decreased protein expression lead to a heterogenous clinical presentation including proximal muscle weakness, contractures, scoliosis, respiratory insufficiency, and ophthalmoplegia. Preclinical model systems of RYR1-RM and MH have been developed to better understand underlying pathomechanisms and test potential therapeutics. Methods: We conducted a comprehensive scoping review of scientific literature pertaining to RYR1-RM and MH preclinical model systems in accordance with the PRISMA Scoping Reviews Checklist and the framework proposed by Arksey and O’Malley. Two major electronic databases (PubMed and EMBASE) were searched without language restriction for articles and abstracts published between January 1, 1990 and July 3, 2019. Results: Our search yielded 5049 publications from which 262 were included in this review. A majority of variants tested in RYR1 preclinical models were localized to established MH/central core disease (MH/CCD) hot spots. A total of 250 unique RYR1 variations were reported in human/rodent/porcine models with 95% being missense substitutions. -
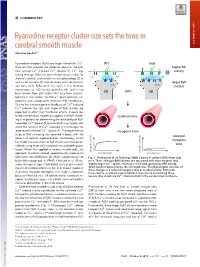
Ryanodine Receptor Cluster Size Sets the Tone in Cerebral Smooth Muscle
COMMENTARY Ryanodine receptor cluster size sets the tone in cerebral smooth muscle COMMENTARY Christian Soellera,1 + Ryanodine receptors (RyRs) are large intracellular Ca2 WT mdx channels that provide the molecular basis of the pro- K+ K+ higher BK + + + cess termed Ca2 -induced Ca2 release (1). Ca2 sig- activity naling through RyRs has been shown to be critical for CaC BK BK skeletal, cardiac, and smooth muscle physiology (2) as well as for neurons (3) and secretory cells like pancre- larger RyR atic beta cells. RyRs were first seen in the electron RyRs RyRs clusters microscope as ∼30-nm-size particles (4), and it had SMCs been known from EM studies that they form clusters, SR SR typically in the narrow “junctional” space between sar- colemma and sarcoplasmic reticulum (SR) membranes. + Due to the inherent positive feedback of Ca2 -induced + Ca2 release, the size and shape of RyR clusters are expected to affect their functional activity. Indeed, de- SMCs SMCs tailed mathematical modeling suggests that RyR cluster- cerebral artery ing is important for determining the excitability of RyR- + mediated Ca2 release (5, 6) and could, in principle, also + affect the amount of Ca2 released in microscopic re- 2+ lease events termed Ca sparks (7). The experimental myogenic tone study of RyR clustering has received a boost with the reduced advent of optical superresolution microscopy, which myogenic has made the assessment of RyR cluster size more acces- sible by using essentially standard immunolabeling pro- tone tocols. When first applied in cardiac muscle cells, this myogenic tone (%) tone myogenic approach revealed a broad, approximately exponential vessel pressure (%) tone myogenic vessel pressure RyR cluster size distribution (8), which had previously not Fig. -

Oxidative Stress Induces Stem Cell Proliferation Via TRPA1/Ryr
RESEARCH ARTICLE Oxidative stress induces stem cell proliferation via TRPA1/RyR-mediated Ca2+ signaling in the Drosophila midgut Chiwei Xu1, Junjie Luo2,3, Li He1, Craig Montell2,3, Norbert Perrimon1,4* 1Department of Genetics, Harvard Medical School, Boston, United States; 2Department of Molecular, Cellular and Developmental Biology, University of California Santa Barbara, Santa Barbara, United States; 3Neuroscience Research Institute, University of California, Santa Barbara, Santa Barbara, United States; 4Howard Hughes Medical Institute, Harvard Medical School, Boston, United States Abstract Precise regulation of stem cell activity is crucial for tissue homeostasis and necessary to prevent overproliferation. In the Drosophila adult gut, high levels of reactive oxygen species (ROS) has been detected with different types of tissue damage, and oxidative stress has been shown to be both necessary and sufficient to trigger intestinal stem cell (ISC) proliferation. However, the connection between oxidative stress and mitogenic signals remains obscure. In a screen for genes required for ISC proliferation in response to oxidative stress, we identified two regulators of cytosolic Ca2+ levels, transient receptor potential A1 (TRPA1) and ryanodine receptor (RyR). Characterization of TRPA1 and RyR demonstrates that Ca2+ signaling is required for oxidative stress-induced activation of the Ras/MAPK pathway, which in turns drives ISC proliferation. Our findings provide a link between redox regulation and Ca2+ signaling and reveal a novel mechanism by which ISCs detect stress signals. DOI: 10.7554/eLife.22441.001 *For correspondence: perrimon@ Introduction receptor.med.harvard.edu Multipotent intestinal stem cells (ISCs) are responsible for tissue homeostasis in the adult Drosophila Competing interests: The midgut (Jiang and Edgar, 2011; Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). -

Original Article Unbalanced Upregulation of Ryanodine Receptor 2 Plays a Particular Role in Early Development of Daunorubicin Cardiomyopathy
Am J Transl Res 2015;7(7):1280-1294 www.ajtr.org /ISSN:1943-8141/AJTR0010909 Original Article Unbalanced upregulation of ryanodine receptor 2 plays a particular role in early development of daunorubicin cardiomyopathy Dana Kucerova1,2, Gabriel Doka1, Peter Kruzliak3, Katarina Turcekova1, Jana Kmecova1, Zuzana Brnoliakova4, Jan Kyselovic1, Uwe Kirchhefer2, Frank U Müller2, Peter Krenek1, Peter Boknik2, Jan Klimas1 1Department of Pharmacology and Toxicology, Faculty of Pharmacy, Comenius University, Bratislava, Slovak Re- public; 2Institut für Pharmakologie und Toxikologie, Universitätsklinikum Münster, Münster, Germany; 3Internation- al Clinical Research Center, St. Anne’s University Hospital and Masaryk University, Brno, Czech Republic; 4Institute of Experimental Pharmacology, Slovak Academy of Sciences, Bratislava, Slovak Republic Received May 31, 2015; Accepted July 14, 2015; Epub July 15, 2015; Published July 30, 2015 Abstract: Calcium release channel on the sarcoplasmic reticulum of cardiomyocytes (ryanodine receptor type 2, RyR2) plays a critical role in the regulation of calcium and was identified as a crucial factor for development of chronic anthracycline cardiomyopathy. Its early stages are less well described although these determine the later development. Hence, we tested the effect of repeated, short-term anthracycline (daunorubicin) administration on cardiac performance, cardiomyocyte function and accompanied changes in calcium regulating proteins expression. Ten-twelve weeks old male Wistar rats were administered with 6 doses of daunorubicin (DAU, 3 mg/kg, i.p., every 48 h), controls (CON) received vehicle. Left ventricular function (left ventricular pressure, LVP; rate of pressure de- velopment, +dP/dt and decline, -dP/dt) was measured using left ventricular catheterization under tribromethanol anaesthesia (15 ml/kg b.w.). -

Alterations of Calcium Homeostasis in Cancer Cells
Available online at www.sciencedirect.com ScienceDirect Alterations of calcium homeostasis in cancer cells Saverio Marchi and Paolo Pinton 2+ Typical hallmarks of cancer include programmed cell death These sources of Ca consist of a large number of 2+ 2+ evasion, uncontrolled cell growth, invasion, and metastasis. Ca pumps, channels, exchangers, and Ca -binding 2+ 2+ Changes in intracellular Ca levels can modulate signaling proteins that aim to control intracellular Ca levels 2+ pathways that control a broad range of cellular events, (Figure 1). Under resting conditions, [Ca ]c is main- including those important to tumorigenesis and cancer tained at a concentration of approximately 100 nM, 2+ progression. Here we discuss how known molecular mediators whereas extracellular [Ca ] is approximately 1 mM. 2+ 2+ of cellular Ca homeostasis impact tumor dynamics and how Ca entry is driven by the presence of a large electro- deregulation of major oncogenes and tumor suppressors is chemical gradient across the plasma membrane. Cells 2+ 2+ tightly associated with Ca signaling. manage this external pool of Ca by activating various Address entry channels with widely different properties. The voltage-gated calcium channels (VGCCs), which belong Department of Morphology, Surgery and Experimental Medicine, Section of Pathology, Oncology and Experimental Biology, Laboratory to the Cav family, are activated by depolarizing mem- for Technologies of Advanced Therapies (LTTA), University of Ferrara, brane potentials and are primarily expressed in excitable -

Novel Types of Ca2+ Release Channels Participate in the Secretory Cycle Of
2 Novel Types of Ca + Release Channels Participate In the Secretory Cycle of Paramecium Cells Eva-Maria Ladenburger,* Ivonne M. Sehring, Iris Korn, and Helmut Plattner Department 0/ Biology, University of Cons/ance, 78457 Constance, Germany A database search of the Paramecium genome reveals 34 genes related to Ca2+ -release channels of the inositol-l,4,S-trisphosphate (IP) or ryanodine receptor type (IP3 R, RyR). Phylogenetic analyses show that these Ca2+ release channels (CRCs) can be subdivided into six groups (Paramecium tetraurelia CRC-I to CRC-VI), each one with features in part reminiscent of 1P3Rs and RyRs. We characterize here the P. tetraurelia CRC-IV-l gene family, whose relationship to IP.,Rs and RyRs is restricted to their C-terminal channel domain. CRC-IV-l channels localize to cortical Ca2+ stores (alveolar sacs) and also to the endoplasmic reticulum. This is in contrast to a recently described true IP3 channel, a group II member (P. tetrallrelia IP3RN -l), found associated with the contractile vacuole system. Silencing of either one of these CRCs results in reduced exocytosis of dense core vesicles (trichocysts), although for different reasons. Knockdown of P. tetraurelia IP3 R N affects trichocyst biogenesis, while CRC-IV-l channels are involved in signal transduction since silenced cells show an impaired release of Ca2+ from cortical stores in response to exocytotic stimuli. Our discovery of a range of CRCs in Paramecium indicates that protozoans already have evolved multiple ways for the use of Ca2+ as signaling molecule. 2 z Ca + is an important component of cell activity in all organ action potentials involving increased [Ca +]; (93). -

Cysteine-Rich Secretory Protein 4 Is an Inhibitor of Transient Receptor Potential M8 with a Role in Establishing Sperm Function
Cysteine-rich secretory protein 4 is an inhibitor of transient receptor potential M8 with a role in establishing sperm function Gerard M. Gibbsa, Gerardo Ortab, Thulasimala Reddya, Adam J. Koppersa, Pablo Martínez-Lópezb, José Luis de la Vega-Beltrànb, Jennifer C. Y. Loa, Nicholas Veldhuisc, Duangporn Jamsaia,d, Peter McIntyrec, Alberto Darszonb,1, and Moira K. O’Bryana,d,1,2 aDepartment of Anatomy and Developmental Biology, and dThe Australian Research Council Centre of Excellence in Biotechnology and Development, Monash University, VIC 3800, Australia; bDepartamento de Genética del Desarrollo y Fisiología Molecular, Instituto de Biotecnología, Universidad Nacional Autónoma de México, Cuernavaca, Morelos, México; and cDepartment of Pharmacology, University of Melbourne, Parkville 3010, Australia Edited by Ryuzo Yanagimachi, The Institute for Biogenesis Research, University of Hawaii, Honolulu, HI, and approved March 18, 2011 (received for review October 28, 2010) The cysteine-rich secretory proteins (CRISPs) are a group of four is the epididymal protein cysteine-rich secretory protein (CRISP) proteins in the mouse that are expressed abundantly in the male 4 (7, 8). reproductive tract, and to a lesser extent in other tissues. Analysis of CRISPs are a subgroup of the CRISP, antigen 5, pathogenesis- reptile CRISPs and mouse CRISP2 has shown that CRISPs can related 1 (CAP) superfamily, which is characterized by the presence of an N-terminal CAP domain (9). CRISPs are verte- regulate cellular homeostasis via ion channels. With the exception fi of the ability of CRISP2 to regulate ryanodine receptors, the in vivo brate-speci c, contain a C-terminal CRISP domain (10, 11), and targets of mammalian CRISPs function are unknown. -

Coming of Age Or Midlife Crisis? Erick O
Hernández-Ochoa and Schneider Skeletal Muscle (2018) 8:22 https://doi.org/10.1186/s13395-018-0167-9 REVIEW Open Access Voltage sensing mechanism in skeletal muscle excitation-contraction coupling: coming of age or midlife crisis? Erick O. Hernández-Ochoa and Martin F. Schneider* Abstract The process by which muscle fiber electrical depolarization is linked to activation of muscle contraction is known as excitation-contraction coupling (ECC). Our understanding of ECC has increased enormously since the early scientific descriptions of the phenomenon of electrical activation of muscle contraction by Galvani that date back to the end of the eighteenth century. Major advances in electrical and optical measurements, including muscle fiber voltage clamp to reveal membrane electrical properties, in conjunction with the development of electron microscopy to unveil structural details provided an elegant view of ECC in skeletal muscle during the last century. This surge of knowledge on structural and biophysical aspects of the skeletal muscle was followed by breakthroughs in biochemistry and molecular biology, which allowed for the isolation, purification, and DNA sequencing of the muscle fiber membrane calcium channel/transverse tubule (TT) membrane voltage sensor (Cav1.1) for ECC and of the muscle ryanodine receptor/sarcoplasmic reticulum Ca2+ release channel (RyR1), two essential players of ECC in skeletal muscle. In regard to the process of voltage sensing for controlling calcium release, numerous studies support the concept that the TT Cav1.1 channel is the voltage sensor for ECC, as well as also being aCa2+ channel in the TT membrane. In this review, we present early and recent findings that support and define the role of Cav1.1 as a voltage sensor for ECC.