University of California, San Diego
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Impact De La Saisonnalité Et D'une Contamination Pesticide
Impact de la saisonnalité et d’une contamination pesticide environnementale sur des relations biotiques entre la micro-méiofaune et les microalgues d’un biofilm d’eau douce Julie Neury-Ormanni To cite this version: Julie Neury-Ormanni. Impact de la saisonnalité et d’une contamination pesticide environnementale sur des relations biotiques entre la micro-méiofaune et les microalgues d’un biofilm d’eau douce. Ecotoxicologie. Université de Bordeaux, 2019. Français. NNT : 2019BORD0343. tel-03091966 HAL Id: tel-03091966 https://tel.archives-ouvertes.fr/tel-03091966 Submitted on 1 Jan 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THÈSE PRÉSENTÉE POUR OBTENIR LE GRADE DE DOCTEUR DE L’UNIVERSITÉ DE BORDEAUX ÉCOLE DOCTORALE : « SCIENCES ET ENVIRONNEMENTS » SPÉCIALITÉ : ECOLOGIE EVOLUTIVE ET FONCTIONNELLE DES COMMUNAUTES Par Julie NEURY-ORMANNI Impact de la saisonnalité et d’une contamination pesticide environnementale sur des relations biotiques entre la micro-méiofaune et les microalgues d’un biofilm d’eau douce Sous la direction de : Soizic MORIN -

Quantum Algorithms for Pattern-Matching in Genomic Sequences
Quantum Algorithms for pattern-matching in genomic sequences Aritra Sarkar Delft University of Technology Quantum Algorithms for pattern-matching in genomic sequences by Aritra Sarkar in partial fulfillment of the requirements for the degree of Master of Science in Computer Engineering at the Delft University of Technology, to be defended publicly on Friday June 22, 2018 at 10:00 AM. Student number: 4597982 Project duration: November 6, 2017 – June 18, 2018 Thesis committee: Prof. dr. ir. Koen Bertels, Q&CE, TU Delft (Supervisor) Dr. ir. Carmen G. Almudever, Q&CE, TU Delft (Daily-supervisor) Dr. Zaid Al-Arz, CE, TU Delft An electronic version of this thesis is available at http://repository.tudelft.nl/. Abstract Fast sequencing and analysis of (microorganism, plant or human) genomes will open up new vistas in fields like personalised medication, food yield and epigenetic research. Current state-of-the-art DNA pattern matching techniques use heuristic algorithms on computing clusters of CPUs, GPUs and FPGAs. With genomic data set to eclipse social and astronomical big data streams within a decade, the alternate computing paradigm of quantum computation is explored to accelerate genome-sequence reconstruction. The inherent parallelism of quantum superposition of states is harnessed to design a quantum kernel for accelerating the search process. The project explores the merger of these two domains and identifies ways to fit these together to design a genome-sequence analysis pipeline with quantum algorithmic speedup. The design of a genome-sequence analysis pipeline with a quantum kernel is tested with a proof-of-concept demonstration using a quantum simulator. -
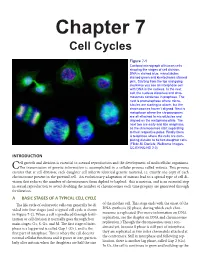
Chapter 7 Cell Cycles
Chapter 7 Cell Cycles Figure 7-1 Confocal micrograph of human cells showing the stages of cell division. DNA is stained blue, microtubules stained green and kinetochores stained pink. Starting from the top and going clockwise you see an interphase cell with DNA in the nucleus. In the next cell, the nucleus dissolves and chro- mosomes condense in prophase. The next is prometaphase where micro- tubules are starting to attach, but the chromosomes haven’t aligned. Next is metaphase where the chromosomes are all attached to microtubules and aligned on the metaphase plate. The next two are early and late anaphase, as the chromosomes start separating to their respective poles. Finally there is telophase where the cells are com- pleting division to be two daughter cells. (Flickr-M. Daniels; Wellcome Images- CC BY-NC-ND 2.0) INTRODUCTION ell growth and division is essential to asexual reproduction and the development of multicellular organisms. CThe transmission of genetic information is accomplished in a cellular process called mitosis. This process ensures that at cell division, each daughter cell inherits identical genetic material, i.e. exactly one copy of each chromosome present in the parental cell. An evolutionary adaptation of mitosis lead to a special type of cell di- vision that reduces the number of chromosomes from diploid to haploid: this is meiosis, and is an essential step in sexual reproduction to avoid doubling the number of chromosomes each time progeny are generated through fertilization. A BASIC STAGES OF A TYPICAL CELL CYCLE The life cycle of eukaryotic cells can generally be di- of the mother cell. -

Améby Skupiny Euamoebida (Amoebozoa, Tubulinea): Vývoj Názorů Na Jejich Taxonomii a Fylogenezi
Jihočeská univerzita v Českých Budějovicích Přírodovědecká fakulta Améby skupiny Euamoebida (Amoebozoa, Tubulinea): Vývoj názorů na jejich taxonomii a fylogenezi Bakalářská práce Simona Školková Školitel: Mgr. Martin Kostka, Ph.D. České Budějovice 2015 Školková, S. (2015): Améby skupiny Euamoebida (Amoebozoa, Tubulinea): Vývoj názorů na jejich taxonomii a fylogenezi. [Amoebas of the group Euamoebida (Amoebozoa, Tubulinea): Development of opinions on the taxonomy and phylogeny. Bc. Thesis, in Czech.] – 44 p., Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic Annotation The study was focused on amoebas of the group Euamoebida (Amoebozoa, Tubulinea). This study attempts to clarify their taxonomy and development of opinions on their phylogeny. It also focuses on changes of naming of individual taxa, which occured during their research. Several other topics (cellular morphology of Amoebozoa, their ecology, characteristics of the families Hartmannellidae and Amoebidae and also relationships between families) are also discussed in this thesis. Prohlašuji, že svoji bakalářskou práci jsem vypracovala samostatně pouze s použitím pramenů a literatury uvedených v seznamu citované literatury. Prohlašuji, že v souladu s § 47b zákona č. 111/1998 Sb. v platném znění souhlasím se zveřejněním své bakalářské práce, a to v nezkrácené podobě elektronickou cestou ve veřejně přístupné části databáze STAG provozované Jihočeskou univerzitou v Českých Budějovicích na jejích internetových stránkách, a to se zachováním mého autorského práva k odevzdanému textu této kvalifikační práce. Souhlasím dále s tím, aby toutéž elektronickou cestou byly v souladu s uvedeným ustanovením zákona č. 111/1998 Sb. zveřejněny posudky školitele a oponentů práce i záznam o průběhu a výsledku obhajoby kvalifikační práce. Rovněž souhlasím s porovnáním textu mé kvalifikační práce s databází kvalifikačních prací Theses.cz provozovanou Národním registrem vysokoškolských kvalifikačních prací a systémem na odhalování plagiátů. -

Identification of Protein Homologous to Inositol Trisphosphate Recep- Tor in Ciliate Blephańsma
NENCKI INSTITUTE OF EXPERIMENTAL BIOLOGY VOLUME 37 NUMBER 4 WARSAWhttp://rcin.org.pl, POLAND 1998 ISSN 0065-1583 Polish Academy of Sciences Nencki Institute of Experimental Biology and Polish Society of Cell Biology ACTA PROTOZOOLOGICA International Journal on Protistology Editor in Chief Jerzy SIKORA Editors Hanna FABCZAK and Anna WASIK Managing Editor Małgorzata WORONOWICZ Editorial Board Andre ADOUTTE, Paris J. I. Ronny LARSSON, Lund Christian F. BARDELE, Tübingen John J. LEE, New York Magdolna Cs. BERECZKY, Göd Jiri LOM, Ćeske Budejovice Y.-Z. CHEN, Beijing Pierangelo LUPORINI, Camerino Jean COHEN, Gif-Sur-Yvette Hans MACHEMER, Bochum John O. COREISS, Albuquerque Jean-Pierre MIGNOT, Aubiere Gyorgy CSABA, Budapest Yutaka NAITOH, Tsukuba Isabelle DESPORTES-LIVAGE, Paris Jytte R. NILSSON, Copenhagen Tom FENCHEL, Helsingor Eduardo ORIAS, Santa Barbara Wilhelm FOISSNER, Salsburg , Dimitrii V. OS SIPO V, St. Petersburg Vassil GOLEMANSKY, Sofia Igor B. RAIKOV, St. Petersburg Andrzej GRĘBECKI, Warszawa, Vice-Chairman Leif RASMUSSEN, Odense Lucyna GRĘBECKA, Warszawa Michael SLEIGH, Southampton Donat-Peter HÄDER, Erlangen Ksenia M. SUKHANOVA, St. Petersburg Janina KACZANOWSKA, Warszawa Jiri VÄVRA, Praha Stanisław L. KAZUBSKI, Warszawa Patricia L. WALNE, Knoxville Leszek KUZNICKI, Warszawa, Chairman ACTA PROTOZOOLOGICA appears quarterly. The price (including Air Mail postage) of subscription to ACTA PROTOZOOLOGICA at 1999 is: US $ 180,- by institutions and US $ 120.- by individual subscribers. Limited number of back volumes at reduced rate are available. TERMS OF PAYMENT: Cheque, money oder or payment to be made to the Nencki Institute of Experimental Biology. Account Number: 11101053-3522-2700-1-34 at Państwowy Bank Kredytowy XIII Oddz. Warszawa, Poland. WITH NOTE: ACTA PROTOZOOLOGICA! For matters regarding ACTA PROTOZOOLOGICA, contact Managing Editor, Nencki Institute of Experimental Biology, ul. -

Analyse De La Variabilité Saisonnière De La Diversité Fonctionnelle Et De La
Analyse de la variabilité saisonnière de la diversité fonctionnelle et de la dynamique des traits liés à l’activité trophique de la microméiofaune dans un biofilm d’eau douce M. Wagner To cite this version: M. Wagner. Analyse de la variabilité saisonnière de la diversité fonctionnelle et de la dynamique des traits liés à l’activité trophique de la microméiofaune dans un biofilm d’eau douce. Sciences de l’environnement. 2018. hal-02607685 HAL Id: hal-02607685 https://hal.inrae.fr/hal-02607685 Submitted on 16 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Analyse de la variabilité saisonnière de la diversité fonctionnelle et de la dynamique des traits liés à l'activité trophique de la microméiofaune dans un biofilm d'eau douce Auteure Margot Wagner CENTRE IRSTEA DE BORDEAUX, UNITE EABX, EQUIPE ECOVEA Encadrante Julie Neury-Ormanni Co-encadrement Jacky Vedrenne et Maud Pierre UNIVERSITE DE RENNES 1 MASTER MENTION BEE . PARCOURS MODELISATION EN ECOLOGIE . PREMIERE ANNEE Référente universitaire Alexandrine Pannard Période de stage : du 3 avril au 31 juillet 2018 Soutenu à Rennes le : 13 juin 2018 INTRODUCTION En écologie, le lien entre réseau trophique et diversité reste une question fondamentale souvent mise en relation avec la structure des communautés biologiques (Paine, 1966), voire de la structure de l’écosystème. -

Genome Projects Have Generally Become Small-Scale Affairs That Genome Are Often Carried out by an Annota�On Individual Laboratory
Genome project Biological significance Sequence Assembly • Genome projects have generally become small-scale affairs that Genome are often carried out by an annotaon individual laboratory. • Genome annotation: – gene prediction & functional Downstream annotation analysis 2 Eukaryo5c genome annotaon Sequencing has become quick and cheap, but annotaon has become more challenging. Shorter read length of NGS The contents of genome are oen terra incognita 6 Genome annotaon 1. General consideraon about gene and genomes 2. Genome Repeat Masking 3. Gene Finding 4. Gene annotaon General Variables of Genomes • Prokaryote versus Eukaryote versus Organelle • Genome size: – Number of chromosomes – Number of base pairs – Number of genes • GC/AT relave content • Repeat content • Genome duplicaons and polyploidy • Gene content See: Genomes, 2nd edion Terence A Brown. ISBN-10: 0-471-25046-5 See NCBI Bookshelve: hVp://www.ncbi.nlm.nih.gov/books/NBK21128/ Eukaryote versus Prokaryote Genomes Eukaryote Prokaryote • Large (10 Mb – 100,000 Mb) • Generally small (<10 Mb; most < 5Mb) • There is not generally a Size relationship between organism • Complexity (as measured by # of genes complexity and its genome size and metabolism) generally proportional (many plants have larger to genome size genomes than human!) Content • Most DNA is non-coding • DNA is “coding gene dense” • Circular DNA, doesn't need telomeres Telomeres/ • Present (Linear DNA) Centromeres • Don’t have mitosis, hence, no centromeres. • More than one, (often) including Number of • Often one, sometimes -

Near-Chromosome Level Genome Assembly Reveals Ploidy Diversity and Plasticity in the Intestinal Protozoan Parasite Entamoeba Histolytica
Kawano-Sugaya et al. BMC Genomics (2020) 21:813 https://doi.org/10.1186/s12864-020-07167-9 RESEARCH ARTICLE Open Access Near-chromosome level genome assembly reveals ploidy diversity and plasticity in the intestinal protozoan parasite Entamoeba histolytica Tetsuro Kawano-Sugaya1,2†, Shinji Izumiyama2†, Yasuaki Yanagawa3, Yumiko Saito-Nakano2, Koji Watanabe3, Seiki Kobayashi4, Kumiko Nakada-Tsukui1,2 and Tomoyoshi Nozaki1* Abstract Background: Amoebozoa is a eukaryotic supergroup composed of unicellular and multicellular amoebic protozoa (e.g. Acanthamoeba, Dictyostelium, and Entamoeba). They are model organisms for studies in cellular and evolutionary biology and are of medical and veterinary importance. Despite their importance, Amoebozoan genome organization and genetic diversity remain poorly studied due to a lack of high-quality reference genomes. The slime mold Dictyostelium discoideum is the only Amoebozoan species whose genome is available at the chromosome-level. Results: Here, we provide a near-chromosome-level assembly of the Entamoeba histolytica genome, the second semi-completed Amoebozoan genome. The availability of this improved genome allowed us to discover inter-strain heterogeneity in ploidy at the near-chromosome or sub-chromosome level among 11 clinical isolates and the reference strain. Furthermore, we observed ploidy-independent regulation of gene expression, contrary to what is observed in other organisms, where RNA levels are affected by ploidy. Conclusions: Our findings offer new insights into Entamoeba chromosome organization, ploidy, transcriptional regulation, and inter-strain variation, which will help to further decipher observed spectrums of virulence, disease symptoms, and drug sensitivity of E. histolytica isolates. Keywords: Entamoeba, Aneuploidy, Expression regulation, Hi-C, PacBio Background environment. Thus, it has evolved unique core metabol- Entamoeba histolytica is an intestinal protozoan parasite ism, mitochondrial structure and function, and cellular that causes human amebiasis. -

Quasi-Species ~~ Evolution at the Speed of Light ~~
Quasi-Species ~~ evolution at the speed of light ~~ Signals and Systems in Biology Kushal Shah @ EE, IIT Delhi Quasi-Species : Introduction I Species : Single genotype I Quasispecies I Large group of genotypes with high mutation rate I RNA viruses, Macromolecules like RNA/DNA I Proposed by Manfred Eigen and Peter Schuster in 1970s J. J. Bull et. al., PLOS Computational Biology 1, e61 (2005) C. O. Wilke, BMC Evolutionary Biology 5:44 (2005) Quasi-Species : Introduction I Species : Single genotype I Quasispecies I Large group of genotypes with high mutation rate I RNA viruses, Macromolecules like RNA/DNA I Proposed by Manfred Eigen and Peter Schuster in 1970s J. J. Bull et. al., PLOS Computational Biology 1, e61 (2005) C. O. Wilke, BMC Evolutionary Biology 5:44 (2005) Quasi-Species : Introduction I Species : Single genotype I Quasispecies I Large group of genotypes with high mutation rate I RNA viruses, Macromolecules like RNA/DNA I Proposed by Manfred Eigen and Peter Schuster in 1970s J. J. Bull et. al., PLOS Computational Biology 1, e61 (2005) C. O. Wilke, BMC Evolutionary Biology 5:44 (2005) Quasi-Species : Introduction I Species : Single genotype I Quasispecies I Large group of genotypes with high mutation rate I RNA viruses, Macromolecules like RNA/DNA I Proposed by Manfred Eigen and Peter Schuster in 1970s J. J. Bull et. al., PLOS Computational Biology 1, e61 (2005) C. O. Wilke, BMC Evolutionary Biology 5:44 (2005) RNA Virus I Contains single- or double-stranded RNA as its genetic material I SARS, influenza, hepatitis C, West Nile -

Bardzo Zróżnicowanych, Ale S³abo Poznanych Eukariotów
PRACE PRZEGL¥DOWE Genomika protistów – bardzo zró¿nicowanych, ale s³abo poznanych eukariotów Pawe³ Mackiewicz1, Przemys³aw Gagat1, Andrzej Body³2 1Zak³ad Genomiki, Wydzia³ Biotechnologii, Uniwersytet Wroc³awski, Wroc³aw 2Zak³ad Bioró¿norodnoœci i Taksonomii Ewolucyjnej, Instytut Zoologiczny, Uniwersytet Wroc³awski, Wroc³aw Genomics of Protists – very diversified but poorly studied eukaryotes Summary Initially, most eukaryotic sequence projects were devoted to typical ani- mals, fungi and plants. Now, more and more effort is put into sequencing pro- tist genomes. Protists are an artificial assemblage that usually contains unicellu- lar eukaryotes coming from different phylogenetic lineages, much more diversi- fied and widespread than higher Eukaryota. The sequenced protist genomes are essential for reconstruction of the Tree of Life and understanding significant events in eukaryotic evolution and diversification. Many protists are parasites and pathogens of medical and economic significance, and play an important ecological role as primary producers and crucial links in food webs. A lot of protists also serve as model organisms in many biological fields and are becom- Adres do korespondencji ing important in biotechnology. 37 protist genome projects were published Pawe³ Mackiewicz, until the beginning of 2010 and 217 are ongoing. Knowledge coming from these Zak³ad Genomiki, projects will be helpful in more efficient protection from pathogenic protists Wydzia³ Biotechnologii, and their elimination. Sequenced genomes of ecologically important protists Uniwersytet Wroc³awski, ul. Przybyszewskiego 63/77, could help to understand many environmental phenomena and even to control 51-148 Wroc³aw; them. Thanks to the newly sequenced genomes we can discover unknown en- e-mail: zymes and metabolic pathways that will be useful in many branches of biotech- [email protected]. -

The Magnitude and Diversity of Infectious Diseases
Chapter 1 The Magnitude and Diversity of Infectious Diseases “All interest in disease and death is only another expression of interest in life.” Thomas Mann THE IMPORTANCE OF INFECTIOUS DISEASES IN TERMS OF HUMAN MORTALITY According to the U.S. Census Bureau, on July 20, 2011, the USA population was 311 806 379, and the world population was 6 950 195 831 [2]. The U.S. Central Intelligence agency estimates that the USA crude death rate is 8.36 per 1000 and the world crude death rate is 8.12 per 1000 [3]. This translates to 2.6 million people dying in 2011 in the USA, and 56.4 million people dying worldwide. These numbers, calculated from authoritative sources, correlate surprisingly well with the widely used rule of thumb that 1% of the human population dies each year. How many of the world’s 56.4 million deaths can be attributed to infectious diseases? According to World Health Organization, in 1996, when the global death toll was 52 million, “Infectious diseases remain the world’s leading cause of death, accounting for at least 17 million (about 33%) of the 52 million people who die each year” [4]. Of course, only a small fraction of infections result in death, and it is impossible to determine the total incidence of infec- tious diseases that occur each year, for all organisms combined. Still, it is useful to consider some of the damage inflicted by just a few of the organisms that infect humans. Malaria infects 500 million people. About 2 million people die each year from malaria [4]. -

Guide to the Methods of Study and Identification of Soil Gymnamoebae
Protistology 3 (3), 148190 (2004) Protistology Guide to the methods of study and identification of soil gymnamoebae Alexey V. Smirnov1 and Susan Brown2 1 Department of Invertebrate Zoology, Faculty of Biology and Soil Sciences, St.Petersburg State University, Russia 2 CEH Dorset, Dorchester, UK Summary The present guide is an attempt to build a “bridge” between the general textbooks on protozoa and the guides to amoebae intended for specialists. We try to outline the subset of freshwater amoebae species that may be found in the soil and list them in the text. The extended introduction section provides detailed descriptions of the methods and shortcomings of amoeba investigation and gives one some ideas on the peculiarities, biology and ecology of soil amoebae. Special section guides the reader through the identification process to prevent him from potential errors. From our experience, dichotomous keys to amoebae are rather artificial and difficult in use, so this guide is based on a classification system of amoeba morphotypes, i.e. on the classification of the generalized shapes of the locomotive form of an amoeba. It allows easy and fast initial classification of an amoeba into one of 16 groups of species containing from two to twentyfive species. The section dedicated to each morphotype contains the sample plate of photographs and the list of relevant literature for further identification of species of the chosen morphotype. Key words: gymnamoebae, amoeba, systematics, identification, methods, guide Foreword morphological and ultrastructural data) requires establishment of cultures and both light (LM) and Amoeboid protists are among the most common electron (EM) microscopy examination, and is highly and abundant microbes in all types of soil habitats.