Mollusca, Gastropoda, Heterobranchia)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

BULLETIN (Mailed to Financial Members of the Society Within Victoria) Price 50¢ EDITOR Val Cram
THE MALACOLOGICAL SOCIETY OF AUSTRALASIA Inc. VICTORIAN BRANCH BULLETIN (Mailed to financial members of the Society within Victoria) Price 50¢ EDITOR Val Cram. Tel. No. 9792 9163 ADDRESS: 6 Southdean Street, Dandenong, Vic. 3175 Conus marmoreus Linne EMAIL: [email protected] VIC. BR. BULL. NO. 269 JUNE/JULY 2013 NOTICE OF MEETING The next meeting of the Branch will be held on the 17th June at the Melbourne Camera Club Building, cnr. Dorcas & Ferrars Sts South Melbourne at 8pm. This will be a Member’s night. Raffles & Supper as usual. There will be no meeting in July. A Bulletin will be issued prior to the August meeting which will be held on the 19th. At the April meeting we welcomed Caitlin Woods, PR Officer for the Malacological Society of Australasia. We discussed with her our role in the society and she offered any assistance she could to promote our branch to further the study of molluscs in Victoria. Jack Austin advises, with considerable regret, that he must dispose of his shell collection as his intended successor-grandson has opted for a volunteer career overseas and will not have a house in Australia for some years. Jack is a part-sponsor of this venture and will sell-off what he can of the collection to raise funds for his grandson. The collection is fairly extensive world-wide, about 7,000 lots, emphasising GBR, SE Australia, NT, Pacific lslands. All lots are registered - lists of families or places can be supplied. Contact details" 11 Station St., Hastings, Vic. (03) 59797242 Secretary/Treasurer Michael Lyons Tel. -

Gyraulus Laevis in Nederland 87
Kuijper: Gyraulus laevis in Nederland 87 Gyraulus laevis (Mollusca: Planorbidae) in Nederland door W.J. Kuijper Enkele recente waarnemingen van een van onze zeldzaamste planorbiden waren de aanleiding tot het samenstellen van een over- laevis zicht van alle van Nederland bekende vondsten van Gyraulus (Alder, 1838). Tot voor kort waren slechts enkele vindplaatsen van dit dier bekend (Janssen & De Vogel, 1965: 75). Hierbij komt het betrof feit dat het in een aantal gevallen slechts een enkel exemplaar en de soort niet meer teruggevonden kon worden. Het volgende geeft een chronologisch overzicht van de waarnemingen van Gyraulus laevis in Nederland. Voor zover beschikbaar zijn diverse gegevens van de vindplaatsen vermeld. WAARNEMINGEN 1. Koudekerke (bij Middelburg), buitenplaats Vijvervreugd, ± 1890. Fraai vers materiaal: 69 exemplaren in de collectie Schep- de man (Zoölogisch Museum, Amsterdam). Later niet meer vermeld, buitenplaats bestaat niet meer (Kuiper, 1944: 6). Dit materiaalwerd verzameld door Dr. IJ. Keijzer en is vermeld in: Verslag over 1885-1893 van het Zeeuwsch Genootschap der Wetenschappen, blz. 88 (volgens Mevr. Dr. W.S.S. van der Feen - van Benthem Jut- ting). Verder is materiaal van deze vindplaats in de collecties van het Zeeuwsch Museum te Middelburg (2 ex.) en het Natuurhistorisch Museum te Enschede (6 ex.) aanwezig. 2. Warmond (bij Leiden), in de Leede, januari 1916 (Van Ben- them Jutting, 1933: 179). Dit materiaal werd verzameld door P.P. de Koning; in het Rijksmuseum van Natuurlijke Historie te Leiden bevinden zich vier schelpen van deze vindplaats, waarvan twee een doorsnede van ca. 4 mm bereiken. 3. 't Zand (bij Roodeschool), Oostpolder, 1927 (Van Benthem Jutting, 1947: 66). -

SCAMIT Newsletter Vol. 22 No. 6 2003 October
October, 2003 SCAMIT Newsletter Vol. 22, No. 6 SUBJECT: B’03 Polychaetes continued - Polycirrus spp, Magelonidae, Lumbrineridae, and Glycera americana/ G. pacifica/G. nana. GUEST SPEAKER: none DATE: 12 Jaunuary 2004 TIME: 9:30 a.m. to 3:30 p. m. LOCATION: LACMNH - Worm Lab SWITCHED AT BIRTH The reader may notice that although this is only the October newsletter, the minutes from the November meeting are included. This is not proof positive that time travel is possible, but reflects the mysterious translocation of minutes from the September meeting to a foster home in Detroit. Since the November minutes were typed and ready to go, rather than delay yet another newsletter during this time of frantic “catching up”, your secretary made the decision to go with what was available. Let me assure everyone that the September minutes will be included in next month’s newsletter. Megan Lilly (CSD) NOVEMBER MINUTES The October SCAMIT meeting on Piromis sp A fide Harris 1985 miscellaneous polychaete issues was cancelled Anterior dorsal view. Image by R. Rowe due to the wildfire situation in Southern City of San Diego California. It has been rescheduled for January ITP Regional 2701 rep. 1, 24July00, depth 264 ft. The SCAMIT Newsletter is not deemed to be a valid publication for formal taxonomic purposes. October, 2003 SCAMIT Newsletter Vol. 22, No. 6 12th. The scheduled topics remain: 1) made to accommodate all expected Polycirrus spp, 2) Magelonidae, 3) participants. If you don’t have his contact Lumbrineridae, and 4) Glycera americana/G. information, RSVP to Secretary Megan Lilly at pacifica/G. -

Nudipleura Bathydorididae Bathydoris Clavigera AY165754 2064 AY427444 1383 AF249222 445 AF249808 599
!"#$"%&'"()*&**'+),#-"',).+%/0+.+()-,)12+),",1+.)$./&3)1/),+-),'&$,)45&("3'+&.-6) !"#$%&'()*"%&+,)-"#."%)-'/%0(%1/'2,3,)45/6"%7/')89:0/5;,)8/'(7")<=)>(5#&%?)@)A(BC"/5)DBC'E752,3 +F/G"':H/%:)&I)A"'(%/)JB&#K#:/H#)FK%"H(B#,)4:H&#GC/'/)"%7)LB/"%)M/#/"'BC)N%#.:$:/,)OC/)P%(Q/'#(:K)&I)O&RK&,)?S+S?) *"#C(T"%&C",)*"#C(T",)UC(V")2WWSX?Y;,)Z"G"%=)2D8D-S-"Q"'("%)D:":/)U&55/B.&%)&I)[&&5&1K,)A9%BCC"$#/%#:'=)2+,)X+2;W) A9%BC/%,)</'H"%K=)3F/G"':H/%:)-(&5&1K)NN,)-(&[/%:'$H,)\$7T(1SA"6(H(5("%#SP%(Q/'#(:]:,)<'&^C"7/'%/'#:'=)2,)X2+?2) _5"%/11SA"'.%#'(/7,)</'H"%K`);D8D-S-"Q"'("%)D:":/)U&55/B.&%)&I)_"5/&%:&5&1K)"%7)</&5&1K,)</&V(&)U/%:/')\AP,) M(BC"'7S>"1%/'SD:'=)+a,)Xa333)A9%BC/%,)</'H"%K`)?>/#:/'%)4$#:'"5("%)A$#/$H,)\&BR/7)-"1);b,)>/5#CG&&5)FU,)_/':C,) >4)YbXY,)4$#:'"5("=))U&''/#G&%7/%B/)"%7)'/c$/#:#)I&')H":/'("5#)#C&$57)V/)"77'/##/7):&)!=*=)d/H"(5e)R"%&f"&'(=$S :&RK&="B=gGh) 7&33'+8+#1-.9)"#:/.8-;/#<) =-*'+)7>?)8$B5/&.7/)#/c$/%B/#)&I)G'(H/'#)$#/7)I&')"HG5(iB".&%)"%7)#/c$/%B(%1 =-*'+)7@?)<"#:'&G&7)#G/B(/#)"%7)#/c$/%B/#)$#/7)(%):C/)GCK5&1/%/.B)'/B&%#:'$B.&%)&I)/$:CK%/$'"%)B5"7/#)(%B5$7(%1) M(%1(B$5&(7/" A"$&.+)7>?)M46A\):'//#)V"#/7)&%)I&$'S1/%/)7":"#/:)T(:C&$:)&%/)&I):T&)H"g&')%$7(G5/$'"%)#$VB5"7/#e)d"h)8$7(V'"%BC(") d!"#$%&'()*+"%7),-.)/)&"h)"%7)dVh)_5/$'&V'"%BC&(7/")d0.-1('2("34$1*+"%7)5'/#$'/6*'3)"h= A"$&.+)7@?)O(H/SB"5(V'":/7)-J4DO):'//#)T(:C&$:)&%/)&I)I&$')B"5(V'".&%)G'(&'#e)d"h)i'#:)#G5(:)T(:C(%)J$&G(#:C&V'"%BC(")"%7) dVh)#G5(:#)V/:T//%)7"(%4$)1/)"%7)8/-"9'.)"%7)dBh)V/:T//%):)39)41.'6*)*)"%7):C'//)&:C/')'(%1(B$5(7#= A"$&.+)7B?)A'-"K/#):'//)V"#/7)&%)I&$'S1/%/)7":"#/:= -
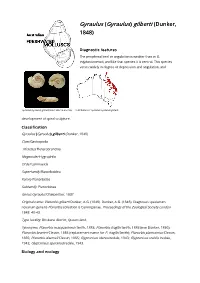
Gyraulus) Gilberti (Dunker, 1848
Gyraulus (Gyraulus) gilberti (Dunker, 1848) Diagnostic features The peripheral keel or angulation is weaker than in G. edgbastonensis, and like that species it is central. This species varies widely in degree of depression and angulation, and Gyraulus (Gyraulus) gilberti (adult size 4.8-5.5 mm) Distribution of Gyraulus (Gyraulus) gilberti. development of spiral sculpture. Classification Gyraulus (Gyraulus) gilberti (Dunker, 1848) Class Gastropoda I nfraclass Heterobranchia Megaorder Hygrophila Order Lymnaeida Superfamily Planorboidea Family Planorbidae Subfamily: Planorbinae Genus Gyraulus Charpentier, 1837 Original name: Planorbis gilberti Dunker, A.G. (1848). Dunker, A.G. (1848). Diagnoses specierum novarum generis Planorbis collection is Cumingianae. Proceedings of the Zoological Society London 1848: 40-43. Type locality: Brisbane district, Queensland. Synonyms: Planorbis macquariensis Smith, 1883; Planorbis fragilis Smith, 1883 (non Dunker, 1850); Planorbis brazieri Clessin, 1885 (replacement name for P. fragilis Smith); Planorbis planissimus Clessin, 1885; Planorbis daemeli Clessin, 1885; Glyptanisus idenusredale, 1943; Glyptanisus stabilis redale, 1943; Glyptanisus speranusredale, 1943. Biology and ecology This species lives in water weeds and other vegetation in ponds, billabongs, swamps and sluggish streams and rivers in tropical and subtropical eastern Australia. Feeds on detritus. Egg mass presumably a jelly strip containing small eggs. Development direct. Brown (2001) described the anatomy of this species. This species is an intermediate host for the stomach fluke Orthocoelium streptocoelium (Boray, 1982; Beesley et al., 1998). Distribution This species occurs throughout eastern Australia, from Cape York to northern New South Wales. Notes G. isingi and/or G.waterhousei may possibly be conspecific with this species (Brown, 2001). Further reading Beesley, P. L., Ross, G. J. -

Anisus Vorticulus (Troschel 1834) (Gastropoda: Planorbidae) in Northeast Germany
JOURNAL OF CONCHOLOGY (2013), VOL.41, NO.3 389 SOME ECOLOGICAL PECULIARITIES OF ANISUS VORTICULUS (TROSCHEL 1834) (GASTROPODA: PLANORBIDAE) IN NORTHEAST GERMANY MICHAEL L. ZETTLER Leibniz Institute for Baltic Sea Research Warnemünde, Seestr. 15, D-18119 Rostock, Germany Abstract During the EU Habitats Directive monitoring between 2008 and 2010 the ecological requirements of the gastropod species Anisus vorticulus (Troschel 1834) were investigated in 24 different waterbodies of northeast Germany. 117 sampling units were analyzed quantitatively. 45 of these units contained living individuals of the target species in abundances between 4 and 616 individuals m-2. More than 25.300 living individuals of accompanying freshwater mollusc species and about 9.400 empty shells were counted and determined to the species level. Altogether 47 species were identified. The benefit of enhanced knowledge on the ecological requirements was gained due to the wide range and high number of sampled habitats with both obviously convenient and inconvenient living conditions for A. vorticulus. In northeast Germany the amphibian zones of sheltered mesotrophic lake shores, swampy (lime) fens and peat holes which are sun exposed and have populations of any Chara species belong to the optimal, continuously and densely colonized biotopes. The cluster analysis emphasized that A. vorticulus was associated with a typical species composition, which can be named as “Anisus-vorticulus-community”. In compliance with that both the frequency of combined occurrence of species and their similarity in relative abundance are important. The following species belong to the “Anisus-vorticulus-community” in northeast Germany: Pisidium obtusale, Pisidium milium, Pisidium pseudosphaerium, Bithynia leachii, Stagnicola palustris, Valvata cristata, Bathyomphalus contortus, Bithynia tentaculata, Anisus vortex, Hippeutis complanatus, Gyraulus crista, Physa fontinalis, Segmentina nitida and Anisus vorticulus. -

“Opisthobranchia”! a Review on the Contribution of Mesopsammic Sea Slugs to Euthyneuran Systematics
Thalassas, 27 (2): 101-112 An International Journal of Marine Sciences BYE BYE “OPISTHOBRANCHIA”! A REVIEW ON THE CONTRIBUTION OF MESOPSAMMIC SEA SLUGS TO EUTHYNEURAN SYSTEMATICS SCHRÖDL M(1), JÖRGER KM(1), KLUSSMANN-KOLB A(2) & WILSON NG(3) Key words: Mollusca, Heterobranchia, morphology, molecular phylogeny, classification, evolution. ABSTRACT of most mesopsammic “opisthobranchs” within a comprehensive euthyneuran taxon set. During the last decades, textbook concepts of “Opisthobranchia” have been challenged by The present study combines our published morphology-based and, more recently, molecular and unpublished topologies, and indicates that studies. It is no longer clear if any precise distinctions monophyletic Rhodopemorpha cluster outside of can be made between major opisthobranch and Euthyneura among shelled basal heterobranchs, acte- pulmonate clades. Worm-shaped, mesopsammic taxa onids are the sister to rissoellids, and Nudipleura such as Acochlidia, Platyhedylidae, Philinoglossidae are the basal offshoot of Euthyneura. Furthermore, and Rhodopemorpha were especially problematic in Pyramidellidae, Sacoglossa and Acochlidia cluster any morphology-based system. Previous molecular within paraphyletic Pulmonata, as sister to remaining phylogenetic studies contained a very limited sampling “opisthobranchs”. Worm-like mesopsammic hetero- of minute and elusive meiofaunal slugs. Our recent branch taxa have clear independent origins and thus multi-locus approaches of mitochondrial COI and 16S their similarities are the result of convergent evolu- rRNA genes and nuclear 18S and 28S rRNA genes tion. Classificatory and evolutionary implications (“standard markers”) thus included representatives from our tree hypothesis are quite dramatic, as shown by some examples, and need to be explored in more detail in future studies. (1) Bavarian State Collection of Zoology. Münchhausenstr. -

Ringiculid Bubble Snails Recovered As the Sister Group to Sea Slugs
www.nature.com/scientificreports OPEN Ringiculid bubble snails recovered as the sister group to sea slugs (Nudipleura) Received: 13 May 2016 Yasunori Kano1, Bastian Brenzinger2,3, Alexander Nützel4, Nerida G. Wilson5 & Accepted: 08 July 2016 Michael Schrödl2,3 Published: 08 August 2016 Euthyneuran gastropods represent one of the most diverse lineages in Mollusca (with over 30,000 species), play significant ecological roles in aquatic and terrestrial environments and affect many aspects of human life. However, our understanding of their evolutionary relationships remains incomplete due to missing data for key phylogenetic lineages. The present study integrates such a neglected, ancient snail family Ringiculidae into a molecular systematics of Euthyneura for the first time, and is supplemented by the first microanatomical data. Surprisingly, both molecular and morphological features present compelling evidence for the common ancestry of ringiculid snails with the highly dissimilar Nudipleura—the most species-rich and well-known taxon of sea slugs (nudibranchs and pleurobranchoids). A new taxon name Ringipleura is proposed here for these long-lost sisters, as one of three major euthyneuran clades with late Palaeozoic origins, along with Acteonacea (Acteonoidea + Rissoelloidea) and Tectipleura (Euopisthobranchia + Panpulmonata). The early Euthyneura are suggested to be at least temporary burrowers with a characteristic ‘bubble’ shell, hypertrophied foot and headshield as exemplified by many extant subtaxa with an infaunal mode of life, while the expansion of the mantle might have triggered the explosive Mesozoic radiation of the clade into diverse ecological niches. The traditional gastropod subclass Euthyneura is a highly diverse clade of snails and slugs with at least 30,000 living species1,2. -

OREGON ESTUARINE INVERTEBRATES an Illustrated Guide to the Common and Important Invertebrate Animals
OREGON ESTUARINE INVERTEBRATES An Illustrated Guide to the Common and Important Invertebrate Animals By Paul Rudy, Jr. Lynn Hay Rudy Oregon Institute of Marine Biology University of Oregon Charleston, Oregon 97420 Contract No. 79-111 Project Officer Jay F. Watson U.S. Fish and Wildlife Service 500 N.E. Multnomah Street Portland, Oregon 97232 Performed for National Coastal Ecosystems Team Office of Biological Services Fish and Wildlife Service U.S. Department of Interior Washington, D.C. 20240 Table of Contents Introduction CNIDARIA Hydrozoa Aequorea aequorea ................................................................ 6 Obelia longissima .................................................................. 8 Polyorchis penicillatus 10 Tubularia crocea ................................................................. 12 Anthozoa Anthopleura artemisia ................................. 14 Anthopleura elegantissima .................................................. 16 Haliplanella luciae .................................................................. 18 Nematostella vectensis ......................................................... 20 Metridium senile .................................................................... 22 NEMERTEA Amphiporus imparispinosus ................................................ 24 Carinoma mutabilis ................................................................ 26 Cerebratulus californiensis .................................................. 28 Lineus ruber ......................................................................... -

Nueva Especie De Rissoella JE Gray, 1847 (Gastropoda: Heterobranchia )
Rev. Acad. Canar. Cienc, XX (Num. 4), 15-17 (2008) (publicado en septiembre de 2009) NUEVA ESPECIE DE Rissoella J. E. GRAY, 1847 (GASTROPODA: HETEROBRANCHIA) DE LA PENINSULA DE GUANAHACABIBES, PINAR DEL RIO, CUBA J. Espinosa & J. Ortea Institute de Oceanologia, Avda. V n° 18406, E. 184 y 186, Playa La Habana. Cuba RESUMEN Descripcion de una nueva especie del genero Rissoella J. E. Gray, 1847, recolectada en el sistema arrecifal de Cabo de San Antonio, peninsula de Guanahacabibes, con datos sobre la coloracion del animal y la estructura de su concha. Palabras clave: MoUusca, Gastropoda, Heterobranchia, Rissoella, especie nueva, Cuba. ABSTRACT A new species of the genus Rissoella J. E. Gray, 1847 collected in the arrecifal sys- tem of San Antonio Cape, peninsula of Guanahacabibes, Cuba is described, with dates of the living animal and shell structure. Key words: MoUusca, Gastropoda, Heterobranchia, Rissoella, new species, Cuba. 1. INTRODUCCION La aportacion mas reciente sobre el genero Rissoella en Cuba y en el Caribe, es la realizada en un trabajo anterior (ORTEA & ESPINOSA [1]) en el que revisamos la taxo- nomia del genero en el Atlantico americano y describimos siete especies nuevas de estos microcaracoles, todas en las costas de Cuba. En dicho trabajo, establecimos dos grupos de especies, siendo el primero de ellos el que reune conchas con el ombligo abierto, sin cordo- nes ni aristas anteriores a el, cuyos animales pueden presentar o no una pigmentacion dife- rencial de la gonada y del digestivo, apreciable en las primeras vueltas de espira por trans- parencia de la concha; a este ultimo grupo es al que pertenece la especie que describimos a continuacion recolectada en el curso de una de las campafias de colecta realizadas en la peninsula de Guanahacabibes. -

In Compliance with Paragraph 20 of the Action Plan, a Directory of Specialists, Laboratories and Organisations Concerned By
EP United Nations Environment Programme UNEP(DEC)/MED WG.268/6 25 Mai 2005 ANGLAIS ORIGINAL: FRANCAIS MEDITERRANEAN ACTION PLAN Seventh Meeting of the Focal Points for SPAs Seville, 31 May to 3rd of June 2005 EVALUATION REPORT ON IMPLEMENTING THE ACTION PLAN FOR THE CONSERVATION OF MARINE VEGETATION IN THE MEDITERRANEAN SEA UNEP RAC/SPA - Tunis, 2005 UNEP(DEC)/MED WG.268/6 Page 1 EVALUATION REPORT ON IMPLEMENTING THE ACTION PLAN FOR THE CONSERVATION OF MARINE VEGETATION IN THE MEDITERRANEAN SEA Introduction In the context of the Mediterranean Action Plan (MAP), the Contracting Parties to the Barcelona Convention adopted at their Eleventh Ordinary Meeting held in Malta in October 1999 an ‘Action Plan for the conservation of marine vegetation in the Mediterranean Sea’ with an accompanying implementation schedule that ended in 2006. The programme of this Action Plan was as follows: it contained 12 actions ordered in response to both the aims articulated in Article 7, and the priorities stated in Article 8. Programme and schedule for implementing the Action Plan for the conservation of marine vegetation in the Mediterranean Sea Action Deadline 1. Ratifying the new SPA Protocol As quickly as possible First symposium before 2. Mediterranean symposium November 2000, then every four years 3. Guidelines for impact studies October 2000 4. First version of the Mediterranean data bank October 2000 5. First issue of the Directory of specialists, laboratories and organisations concerned by October 2000 marine vegetation in the Mediterranean 6. Launching procedures for the legal protection of Some time in 2001 species at national level 7. -

Guide to the Systematic Distribution of Mollusca in the British Museum
PRESENTED ^l)c trustee*. THE BRITISH MUSEUM. California Swcademu 01 \scienceb RECEIVED BY GIFT FROM -fitoZa£du^4S*&22& fo<?as7u> #yjy GUIDE TO THK SYSTEMATIC DISTRIBUTION OK MOLLUSCA IN III K BRITISH MUSEUM PART I HY JOHN EDWARD GRAY, PHD., F.R.S., P.L.S., P.Z.S. Ac. LONDON: PRINTED BY ORDER OF THE TRUSTEES 1857. PRINTED BY TAYLOR AND FRANCIS, RED LION COURT, FLEET STREET. PREFACE The object of the present Work is to explain the manner in which the Collection of Mollusca and their shells is arranged in the British Museum, and especially to give a short account of the chief characters, derived from the animals, by which they are dis- tributed, and which it is impossible to exhibit in the Collection. The figures referred to after the names of the species, under the genera, are those given in " The Figures of Molluscous Animals, for the Use of Students, by Maria Emma Gray, 3 vols. 8vo, 1850 to 1854 ;" or when the species has been figured since the appear- ance of that work, in the original authority quoted. The concluding Part is in hand, and it is hoped will shortly appear. JOHN EDWARD GRAY. Dec. 10, 1856. ERRATA AND CORRIGENDA. Page 43. Verenad.e.—This family is to be erased, as the animal is like Tricho- tropis. I was misled by the incorrectness of the description and figure. Page 63. Tylodinad^e.— This family is to be removed to PleurobrancMata at page 203 ; a specimen of the animal and shell having since come into my possession.