Orthogonal Ubiquitin Transfer Identifies Ubiquitination Substrates Under
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Organ Level Protein Networks As a Reference for the Host Effects of the Microbiome
Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press 1 Organ level protein networks as a reference for the host effects of the microbiome 2 3 Robert H. Millsa,b,c,d, Jacob M. Wozniaka,b, Alison Vrbanacc, Anaamika Campeaua,b, Benoit 4 Chassainge,f,g,h, Andrew Gewirtze, Rob Knightc,d, and David J. Gonzaleza,b,d,# 5 6 a Department of Pharmacology, University of California, San Diego, California, USA 7 b Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, 8 California, USA 9 c Department of Pediatrics, and Department of Computer Science and Engineering, University of 10 California, San Diego California, USA 11 d Center for Microbiome Innovation, University of California, San Diego, California, USA 12 e Center for Inflammation, Immunity and Infection, Institute for Biomedical Sciences, Georgia State 13 University, Atlanta, GA, USA 14 f Neuroscience Institute, Georgia State University, Atlanta, GA, USA 15 g INSERM, U1016, Paris, France. 16 h Université de Paris, Paris, France. 17 18 Key words: Microbiota, Tandem Mass Tags, Organ Proteomics, Gnotobiotic Mice, Germ-free Mice, 19 Protein Networks, Proteomics 20 21 # Address Correspondence to: 22 David J. Gonzalez, PhD 23 Department of Pharmacology and Pharmacy 24 University of California, San Diego 25 La Jolla, CA 92093 26 E-mail: [email protected] 27 Phone: 858-822-1218 28 1 Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press 29 Abstract 30 Connections between the microbiome and health are rapidly emerging in a wide range of 31 diseases. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

Anti-UBE2Z Monoclonal Antibody (DCABH-13898) This Product Is for Research Use Only and Is Not Intended for Diagnostic Use
Anti-UBE2Z monoclonal antibody (DCABH-13898) This product is for research use only and is not intended for diagnostic use. PRODUCT INFORMATION Antigen Description Modification of proteins with ubiquitin (UBB; MIM 191339) or ubiquitin-like proteins controls many signaling networks and requires a ubiquitin activating enzyme (E1), a ubiquitin conjugating enzyme (E2), and a ubiquitin protein ligase (E3). UBE2Z is an E2 enzyme (Jin et al., 2007 [PubMed 17597759]). Immunogen A synthetic peptide of human UBE2Z is used for rabbit immunization. Isotype IgG Source/Host Rabbit Species Reactivity Human Purification Protein A Conjugate Unconjugated Applications Western Blot (Transfected lysate); ELISA Size 1 ea Buffer In 1x PBS, pH 7.4 Preservative None Storage Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. GENE INFORMATION Gene Name UBE2Z ubiquitin-conjugating enzyme E2Z [ Homo sapiens ] Official Symbol UBE2Z Synonyms UBE2Z; ubiquitin-conjugating enzyme E2Z; ubiquitin-conjugating enzyme E2 Z; FLJ13855; UBA6 specific enzyme E2; USE1; UBA6-specific enzyme E2; ubiquitin-protein ligase Z; ubiquitin carrier protein Z; uba6-specific E2 conjugating enzyme 1; HOYS7; Entrez Gene ID 65264 Protein Refseq NP_075567 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 1 © Creative Diagnostics All Rights Reserved UniProt ID Q9H832 Chromosome Location 17q21.32 Pathway Adaptive Immune System, organism-specific biosystem; Antigen processing: Ubiquitination & Proteasome degradation, organism-specific biosystem; Class I MHC mediated antigen processing & presentation, organism-specific biosystem; Immune System, organism-specific biosystem; Ubiquitin mediated proteolysis, organism-specific biosystem; Function ATP binding; acid-amino acid ligase activity; ligase activity; nucleotide binding; ubiquitin-protein ligase activity; 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 2 © Creative Diagnostics All Rights Reserved. -

Supporting Information
Supporting Information Pouryahya et al. SI Text Table S1 presents genes with the highest absolute value of Ricci curvature. We expect these genes to have significant contribution to the network’s robustness. Notably, the top two genes are TP53 (tumor protein 53) and YWHAG gene. TP53, also known as p53, it is a well known tumor suppressor gene known as the "guardian of the genome“ given the essential role it plays in genetic stability and prevention of cancer formation (1, 2). Mutations in this gene play a role in all stages of malignant transformation including tumor initiation, promotion, aggressiveness, and metastasis (3). Mutations of this gene are present in more than 50% of human cancers, making it the most common genetic event in human cancer (4, 5). Namely, p53 mutations play roles in leukemia, breast cancer, CNS cancers, and lung cancers, among many others (6–9). The YWHAG gene encodes the 14-3-3 protein gamma, a member of the 14-3-3 family proteins which are involved in many biological processes including signal transduction regulation, cell cycle pro- gression, apoptosis, cell adhesion and migration (10, 11). Notably, increased expression of 14-3-3 family proteins, including protein gamma, have been observed in a number of human cancers including lung and colorectal cancers, among others, suggesting a potential role as tumor oncogenes (12, 13). Furthermore, there is evidence that loss Fig. S1. The histogram of scalar Ricci curvature of 8240 genes. Most of the genes have negative scalar Ricci curvature (75%). TP53 and YWHAG have notably low of p53 function may result in upregulation of 14-3-3γ in lung cancer Ricci curvatures. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Arsenic Hexoxide Has Differential Effects on Cell Proliferation And
www.nature.com/scientificreports OPEN Arsenic hexoxide has diferential efects on cell proliferation and genome‑wide gene expression in human primary mammary epithelial and MCF7 cells Donguk Kim1,7, Na Yeon Park2,7, Keunsoo Kang3, Stuart K. Calderwood4, Dong‑Hyung Cho2, Ill Ju Bae5* & Heeyoun Bunch1,6* Arsenic is reportedly a biphasic inorganic compound for its toxicity and anticancer efects in humans. Recent studies have shown that certain arsenic compounds including arsenic hexoxide (AS4O6; hereafter, AS6) induce programmed cell death and cell cycle arrest in human cancer cells and murine cancer models. However, the mechanisms by which AS6 suppresses cancer cells are incompletely understood. In this study, we report the mechanisms of AS6 through transcriptome analyses. In particular, the cytotoxicity and global gene expression regulation by AS6 were compared in human normal and cancer breast epithelial cells. Using RNA‑sequencing and bioinformatics analyses, diferentially expressed genes in signifcantly afected biological pathways in these cell types were validated by real‑time quantitative polymerase chain reaction and immunoblotting assays. Our data show markedly diferential efects of AS6 on cytotoxicity and gene expression in human mammary epithelial normal cells (HUMEC) and Michigan Cancer Foundation 7 (MCF7), a human mammary epithelial cancer cell line. AS6 selectively arrests cell growth and induces cell death in MCF7 cells without afecting the growth of HUMEC in a dose‑dependent manner. AS6 alters the transcription of a large number of genes in MCF7 cells, but much fewer genes in HUMEC. Importantly, we found that the cell proliferation, cell cycle, and DNA repair pathways are signifcantly suppressed whereas cellular stress response and apoptotic pathways increase in AS6‑treated MCF7 cells. -

Human Use1/UBE2Z Antibody Antigen Affinity-Purified Polyclonal Sheep Igg Catalog Number: AF8154
Human Use1/UBE2Z Antibody Antigen Affinity-purified Polyclonal Sheep IgG Catalog Number: AF8154 DESCRIPTION Species Reactivity Human Specificity Detects human USE1/UBE2Z in direct ELISAs and Western blots. Source Polyclonal Sheep IgG Purification Antigen Affinitypurified Immunogen E. coliderived recombinant human Use1/UBE2Z Met1Val354 Accession # Q9H832 Formulation Lyophilized from a 0.2 μm filtered solution in PBS with Trehalose. See Certificate of Analysis for details. *Small pack size (SP) is supplied either lyophilized or as a 0.2 μm filtered solution in PBS. APPLICATIONS Please Note: Optimal dilutions should be determined by each laboratory for each application. General Protocols are available in the Technical Information section on our website. Recommended Sample Concentration Western Blot 0.5 µg/mL See Below DATA Western Blot Detection of Human Use1/UBE2Z by Western Blot. Western blot shows lysates of A549 human lung carcinoma cell line and PANC1 human pancreatic carcinoma cell line. PVDF membrane was probed with 0.5 µg/mL of Sheep AntiHuman Use1/UBE2Z Antigen Affinitypurified Polyclonal Antibody (Catalog # AF8154) followed by HRPconjugated AntiSheep IgG Secondary Antibody (Catalog # HAF016). A specific band was detected for Use1/UBE2Z at approximately 3941 kDa (as indicated). This experiment was conducted under reducing conditions and using Immunoblot Buffer Group 1. PREPARATION AND STORAGE Reconstitution Reconstitute at 0.2 mg/mL in sterile PBS. Shipping The product is shipped at ambient temperature. Upon receipt, store it immediately at the temperature recommended below. *Small pack size (SP) is shipped with polar packs. Upon receipt, store it immediately at 20 to 70 °C Stability & Storage Use a manual defrost freezer and avoid repeated freezethaw cycles. -
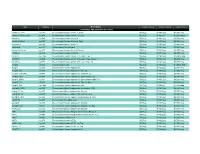
Prospec (Product
Name Catalog # Description Customer Price-A Customer Price-B Customer Price-C CYTOKINES AND GROWTH FACTORS Activin A, Active cyt-145 Recombinant Human Activin-A, Active $50/2µg $130/10µg $3,500/1mg Activin A, Plant-Active cyt-414 Recombinant Human Activin-A Active $50/1µg $130/5µg $1,500/100µg Activin A cyt-569 Recombinant Human Activin-A $50/2µg $130/10µg $2,700/1mg Activin A, Plant cyt-052 Recombinant Human Activin-A, Plant $50/2µg $130/10µg $4,800/1mg mActivin A cyt-146 Recombinant Mouse Activin-A $50/2µg $130/10µg $3,500/1mg rActivin A cyt-147 Recombinant Rat Activin-A $50/2µg $130/10µg $3,500/1mg Activin B, Active cyt-057 Recombinant Human Activin-B Active $50/2µg $130/10µg $5,200/1mg Activin B cyt-058 Recombinant Human Activin-B $50/2µg $130/10µg $4,800/1mg ACVR1 cyt-1140 Recombinant Human Activin A Receptor Type 1 $50/2µg $130/10µg $1,000/0.1mg ACVRL1 cyt-920 Recombinant Human Activin A Receptor Type II-Like 1 $50/2µg $130/10µg $5,200/1mg ACVR2A cyt-976 Recombinant Human Activin A Receptor Type 2A $50/2µg $130/10µg $5,200/1mg Acrp30 cyt-024 Human Adiponectin $50/2µg $130/10µg $1,000/0.1mg Acrp30 cyt-280 Recombinant Human Adiponectin $50/5µg $130/25µg $2,700/1mg Acrp30, His cyt-433 Recombinant Human Adiponectin, His tag $50/10µg $130/50µg $1,900/1mg Acrp30 (108-244) cyt-073 Recombinant Human Adiponectin (108-244 a.a.) $50/5µg $130/25µg $2,700/1mg Acrp30, HEK cyt-434 Recombinant Human Adiponectin glycosilated, HEK $50/2µg $130/10µg $4,680/1mg Acrp30, HMW cyt-764 Recombinant Human Adiponectin glycosilated, HMW Rich $50/2µg $130/10µg -

Cascade Profiling of the Ubiquitin-Proteasome System in Cancer Anastasiia Rulina
Cascade profiling of the ubiquitin-proteasome system in cancer Anastasiia Rulina To cite this version: Anastasiia Rulina. Cascade profiling of the ubiquitin-proteasome system in cancer. Agricultural sciences. Université Grenoble Alpes, 2015. English. NNT : 2015GREAV028. tel-01321321 HAL Id: tel-01321321 https://tel.archives-ouvertes.fr/tel-01321321 Submitted on 25 May 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THÈSE Pour obtenir le grade de DOCTEUR DE L’UNIVERSITÉ GRENOBLE ALPES Spécialité : Biodiversite du Developpement Oncogenese Arrêté ministériel : 7 août 2006 Présentée par Anastasiia Rulina Thèse dirigée par Maxim Balakirev préparée au sein du Laboratoire BIOMICS dans l'École Doctorale Chimie et Sciences du Vivant Profilage en cascade du système ubiquitine- protéasome dans le cancer Thèse soutenue publiquement le «17/12/2015», devant le jury composé de : M. Damien ARNOULT Docteur CR1, CNRS, Rapporteur M. Matthias NEES Professor Adjunct, University of Turku, Rapporteur M. Philippe SOUBEYRAN Docteur, INSERM, Membre Mme. Jadwiga CHROBOCZEK Directeur de recherche DR1, CNRS, Membre M. Xavier GIDROL Directeur de laboratoire, CEA, Président du jury M. Maxim BALAKIREV Docteur, CEA, Membre, Directeur de Thèse 2 Я посвящаю эту научную работу моей маме, Рулиной Людмиле Михайловне, с любовью и благодарностью. -

Table SI. Enriched Genes in the Upregulated Genes of the Recovery Group According to the GO Molecular Function Terms. A, Downreg
Table SI. Enriched genes in the upregulated genes of the recovery group according to the GO Molecular Function terms. A, Downregulated genes Adjusted Total Molecular Rank P‑value genes (n) Function Genes 1 <0.001 266 GO:0019899 Raf1 Timp1 Tbc1d8 Ube2g2 Ube2z enzyme binding Lonrf3 Tbc1d15 Rnf144a Ube2g1 Shc3 Rgcc Rnf19a Ube2j2 Rnf138 Atg13 Cks1b Ube2j1 Rnf19b Trib1 Trib3 Abtb2 Rnf125 Cdc42ep3 Nploc4 Cdc42ep4 Cdc42ep2 Rab11fip5 Arih2 Brms1 Tmem189 Mef2d Hspb1 Cdk9 Ksr1 Tnfaip3 Net1 Rnf180 Fgr Bhlhe41 Irs2 Ppp1r15a Asb4 Trim72 Zfp36 Sfn Xpo6 Fap Sox9 Mapk7 Itga3 Tubb5 Daxx Klf4 Stat3 Gab2 Myo9b Cstb Hmox1 Por Bcl2l1 Plin5 Chp1 Ube2i Sash1 Sqstm1 Rxra Slpi Sdc4 Tnfaip1 Cd40 Slc12a4 Map2k3 Ywhah Ppp1r12a Cry1 Plek Egfr Tnip1 Npc1l1 Rock2 Map2k6 Per1 Nfkbia Bdkrb2 Prkch Hif1a Golga5 Ripk1 Map3k1 Glud1 Nufip1 Clu Spry2 Hcls1 Ifnar2 Tuba1b Cdkn1a Sik1 Tmem173 Map3k2 Tnf Riok3 Ptpn2 Cep192 Smad2 Fas Jak2 Ankrd1 Rela Rps6ka4 Ankrd2 Rabgef1 Prkar1b Nop58 Casp8 Cflar Hdac4 Sele Nek2 Optn Nek6 Lcn2 Stom Traf6 Spred1 Nop56 Src Ccnl1 Ptpn22 Il6ra Pip5k1a F3 Bcl10 3110043O21Rik Tnfrsf1b Slc2a1 Sfpq Rpa2 Errfi1 Mad2l2 Tbc1d14 Uchl1 Glmn Scarb2 Ulk1 Ung Rad18 Mef2a Ctsc Ipo5 Mvp Kctd13 Msn Eif4ebp1 Casp3 Smad1 Ubash3b Ets1 Tirap Smad3 Tgfbr2 Ptgs2 Prr5l Micall1 Cnppd1 Map2k4 Tnks1bp1 Ppp1r32 Prdm4 Midn Ibtk Rusc2 Fmnl2 Ptpn23 Sh3bp4 Nop14 Kdm1a Serpine1 Gch1 Inf2 Csf3 Snx10 Txnip Egr1 Ranbp9 Akap12 Rab3gap2 Ddx58 Bcor Rabggta Pik3r1 Pkp2 Usp22 Shc1 Ptpn11 Fzd5 Cxcr4 Plaur Bag5 Maml1 Camk2n2 Taf7 Ywhag Ezr Jun Camk2d Parp4 Nod2 Ptafr Hmga2 Zfp746 Ptk2b Flot1 -

Discerning the Role of Foxa1 in Mammary Gland
DISCERNING THE ROLE OF FOXA1 IN MAMMARY GLAND DEVELOPMENT AND BREAST CANCER by GINA MARIE BERNARDO Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Dissertation Adviser: Dr. Ruth A. Keri Department of Pharmacology CASE WESTERN RESERVE UNIVERSITY January, 2012 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of Gina M. Bernardo ______________________________________________________ Ph.D. candidate for the ________________________________degree *. Monica Montano, Ph.D. (signed)_______________________________________________ (chair of the committee) Richard Hanson, Ph.D. ________________________________________________ Mark Jackson, Ph.D. ________________________________________________ Noa Noy, Ph.D. ________________________________________________ Ruth Keri, Ph.D. ________________________________________________ ________________________________________________ July 29, 2011 (date) _______________________ *We also certify that written approval has been obtained for any proprietary material contained therein. DEDICATION To my parents, I will forever be indebted. iii TABLE OF CONTENTS Signature Page ii Dedication iii Table of Contents iv List of Tables vii List of Figures ix Acknowledgements xi List of Abbreviations xiii Abstract 1 Chapter 1 Introduction 3 1.1 The FOXA family of transcription factors 3 1.2 The nuclear receptor superfamily 6 1.2.1 The androgen receptor 1.2.2 The estrogen receptor 1.3 FOXA1 in development 13 1.3.1 Pancreas and Kidney