Highlight: Big Surprises from the World’S Smallest Fish
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Resolving Cypriniformes Relationships Using an Anchored Enrichment Approach Carla C
Stout et al. BMC Evolutionary Biology (2016) 16:244 DOI 10.1186/s12862-016-0819-5 RESEARCH ARTICLE Open Access Resolving Cypriniformes relationships using an anchored enrichment approach Carla C. Stout1*†, Milton Tan1†, Alan R. Lemmon2, Emily Moriarty Lemmon3 and Jonathan W. Armbruster1 Abstract Background: Cypriniformes (minnows, carps, loaches, and suckers) is the largest group of freshwater fishes in the world (~4300 described species). Despite much attention, previous attempts to elucidate relationships using molecular and morphological characters have been incongruent. In this study we present the first phylogenomic analysis using anchored hybrid enrichment for 172 taxa to represent the order (plus three out-group taxa), which is the largest dataset for the order to date (219 loci, 315,288 bp, average locus length of 1011 bp). Results: Concatenation analysis establishes a robust tree with 97 % of nodes at 100 % bootstrap support. Species tree analysis was highly congruent with the concatenation analysis with only two major differences: monophyly of Cobitoidei and placement of Danionidae. Conclusions: Most major clades obtained in prior molecular studies were validated as monophyletic, and we provide robust resolution for the relationships among these clades for the first time. These relationships can be used as a framework for addressing a variety of evolutionary questions (e.g. phylogeography, polyploidization, diversification, trait evolution, comparative genomics) for which Cypriniformes is ideally suited. Keywords: Fish, High-throughput -

BMC Evolutionary Biology Biomed Central
BMC Evolutionary Biology BioMed Central Research article Open Access Evolution of miniaturization and the phylogenetic position of Paedocypris, comprising the world's smallest vertebrate Lukas Rüber*1, Maurice Kottelat2, Heok Hui Tan3, Peter KL Ng3 and Ralf Britz1 Address: 1Department of Zoology, The Natural History Museum, Cromwell Road, London SW7 5BD, UK, 2Route de la Baroche 12, Case postale 57, CH-2952 Cornol, Switzerland (permanent address) and Raffles Museum of Biodiversity Research, National University of Singapore, Kent Ridge, Singapore 119260 and 3Department of Biological Sciences, National University of Singapore, Kent Ridge, Singapore 119260 Email: Lukas Rüber* - [email protected]; Maurice Kottelat - [email protected]; Heok Hui Tan - [email protected]; Peter KL Ng - [email protected]; Ralf Britz - [email protected] * Corresponding author Published: 13 March 2007 Received: 23 October 2006 Accepted: 13 March 2007 BMC Evolutionary Biology 2007, 7:38 doi:10.1186/1471-2148-7-38 This article is available from: http://www.biomedcentral.com/1471-2148/7/38 © 2007 Rüber et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Paedocypris, a highly developmentally truncated fish from peat swamp forests in Southeast Asia, comprises the world's smallest vertebrate. Although clearly a cyprinid fish, a hypothesis about its phylogenetic position among the subfamilies of this largest teleost family, with over 2400 species, does not exist. -

Making a Big Splash with Louisiana Fishes
Making a Big Splash with Louisiana Fishes Written and Designed by Prosanta Chakrabarty, Ph.D., Sophie Warny, Ph.D., and Valerie Derouen LSU Museum of Natural Science To those young people still discovering their love of nature... Note to parents, teachers, instructors, activity coordinators and to all the fishermen in us: This book is a companion piece to Making a Big Splash with Louisiana Fishes, an exhibit at Louisiana State Universi- ty’s Museum of Natural Science (MNS). Located in Foster Hall on the main campus of LSU, this exhibit created in 2012 contains many of the elements discussed in this book. The MNS exhibit hall is open weekdays, from 8 am to 4 pm, when the LSU campus is open. The MNS visits are free of charge, but call our main office at 225-578-2855 to schedule a visit if your group includes 10 or more students. Of course the book can also be enjoyed on its own and we hope that you will enjoy it on your own or with your children or students. The book and exhibit was funded by the Louisiana Board Of Regents, Traditional Enhancement Grant - Education: Mak- ing a Big Splash with Louisiana Fishes: A Three-tiered Education Program and Museum Exhibit. Funding was obtained by LSUMNS Curators’ Sophie Warny and Prosanta Chakrabarty who designed the exhibit with Southwest Museum Services who built it in 2012. The oarfish in the exhibit was created by Carolyn Thome of the Smithsonian, and images exhibited here are from Curator Chakrabarty unless noted elsewhere (see Appendix II). -

Cypriniformes of Borneo (Actinopterygii, Otophysi): an Extraordinary Fauna for Integrated Studies on Diversity, Systematics, Evolution, Ecology, and Conservation
Zootaxa 3586: 359–376 (2012) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ ZOOTAXA Copyright © 2012 · Magnolia Press Article ISSN 1175-5334 (online edition) urn:lsid:zoobank.org:pub:7A06704C-8DE5-4B9F-9F4B-42F7C6C9B32F Cypriniformes of Borneo (Actinopterygii, Otophysi): An Extraordinary Fauna for Integrated Studies on Diversity, Systematics, Evolution, Ecology, and Conservation ZOHRAH H. SULAIMAN1 & R.L MAYDEN2 1Biological Science Programme, Faculty of Science, Universiti Brunei Darussalam, Tungku BE1410, Brunei Darussalam; E-mail:[email protected] 2Department of Biology, 3507 Laclede Ave, Saint Louis University, St Louis, Missouri 63103, USA; E-mail:[email protected] Abstract Borneo Island is governed by the countries of Brunei Darussalam, Malaysia (Sabah and Sarawak) and Indonesia (Kalimantan) and is part of Sundaland. These countries have a high diversity of freshwater fishes, especially described and undescribed species of Cypriniformes; together these species and other flora and fauna represent an extraordinary opportunity for worldwide collaboration to investigate the biodiversity, conservation, management and evolution of Borneo’s wildlife. Much of the fauna and flora of Borneo is under significant threat, warranting an immediate and swift international collaboration to rapidly inventory, describe, and conserve the diversity. The Sunda drainage appears to have been an important evolutionary centre for many fish groups, including cypriniforms (Cyprinidae, Balitoridae and Gyrinocheilidae); however, Northwestern Borneo (Brunei, Sabah and Sarawak) is not connected to Sundaland, and this disjunction likely explains the non-homogeneity of Bornean ichthyofauna. A previous study confirmed that northern Borneo, eastern Borneo and Sarawak shared a similar ichthyofauna, findings that support the general hypothesis for freshwater connections at one time between western Borneo and central Sumatra, and south Borneo and Java island. -
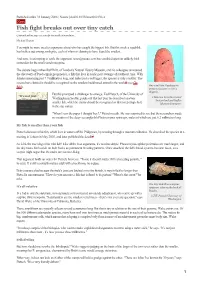
Fish Fight Breaks out Over Tiny Catch
Published online 31 January 2006 | Nature | doi:10.1038/news060130-4 News Fish fight breaks out over tiny catch Contenders line up to net credit for smallest vertebrate. Michael Hopkin You might be more used to arguments about who has caught the biggest fish. But this week a squabble has broken out among zoologists, each of whom is claiming to have found the smallest. And now, in attempting to settle the argument, [email protected] has stumbled upon an unlikely third contender for the small-vertebrate prize. The debate began when Ralf Britz, of London's Natural History Museum, and his colleagues announced the discovery of Paedocypris progenetica, a fish that lives in acidic peat swamps of southeast Asia. With females measuring just 7.9 millimetres long, and males just a tad bigger, the species is truly a tiddler. The researchers claimed it should be recognized as the smallest backboned animal in the world1 (see Go One small fish: Paedocypris fish!). progenetica gets lost on a fingertip. But this prompted a challenger to emerge. Ted Pietsch, of the University of “It's not just Washington in Seattle, points out that last year he described an even © Maurice Kottelat Cornol millimetres that Switzerland and Raffles count - it's how smaller fish, which he claims should be recognized as first (or perhaps last) Museum Singapore you use those in the size stakes. millimetres.” "When I saw the paper I thought 'hey!'," Pietsch recalls. He was surprised to see that the researchers made no mention of the deep-sea anglerfish Photocorynus spiniceps, males of which are just 6.2 millimetres long. -

Betta (2006) NB: 3 of These Species Were Identified from Within HOB
The Search Continues… The Latest Rem arkable Species Discoveries from Borneo July 2005 to Septem ber 2006 This document provides details of species identified as being described from Borneo during the period July 2005 to September 2006. This list has been compiled through web-based research and correspondence with the academic professionals listed, and should not be treated as an exhaustive or comprehensive list of newly discovered species on Borneo. Species discovered within the Heart of Borneo Boundary 01 A new species of tree frog Polypedates chlorophthalmus (December 2005) Name: Polypedates chlorophthalmus (Anura:Rhacophoridae) Description: A new species of treefrog, measuring 62.1 mm long. It has a rounded snout, bright green eyes and a thin dark gray line at back of its forehead. The species is named for its strikingly coloured eyes, its name coming from the Greek for ‘green eyed’. Location: Discovered within the Heart of Borneo boundary, at Gunung Murid Sarawak, the highest mountain in the state of Sarawak, Northwestern Borneo. The genus Polypedates is known to contain 16 nominal species, of which 10 occur in southeast Asia. The members of the genus are distributed from southern China, Sri Lanka and south-western and north-eastern India south to Indo-China and Indo-Malaya (Frost, 1985). Of these, four species have been reported from Borneo (Inger & Stuebing, 1997; Inger & Tan, 1996). Source(s): The Raffles Bulletin of Zoology 2005 53(2): Pp. 265-270. National University of Singapore. Date of Publication: 31 Dec. 2005 Contact: Dr Indraneil Das, Institute of Biodiversity and Environmental Conservation, University of Malaysia: [email protected] The newly discovered species of Borneo Christian Thompson – 17 November 2006 1 02 A new species of catfish Glyptothorax exodon (December 2005) Name: Glyptothorax exodon, (Teleostei: Sisoridae). -

Unique Southeast Asian Peat Swamp Forest Habitats Have Relatively Few Distinctive Plant Species
Unique Southeast Asian peat swamp forest habitats have relatively few distinctive plant species W. Giesen1, L.S. Wijedasa2 and S.E. Page3 1Euroconsult Mott MacDonald, Arnhem, The Netherlands 2Theoretical Ecology & Modelling Laboratory, National University of Singapore and ConservationLinks, Singapore 3Department of Geography, University of Leicester, UK _______________________________________________________________________________________ SUMMARY The peat swamp forests of Southeast Asia are often described as having a unique biodiversity. While these waterlogged and nutrient-poor habitats are indeed unique and include a distinct fauna (especially fish), the peat swamp forest flora is much less distinct and shares a surprisingly large number of species with other habitats. Out of 1,441 species of higher plants found in Southeast Asian swamps (from Thailand to Papua), 1,337 are found in the lowlands (< 300 m a.s.l.). Of these 1,337 species, 216 (16.2 %) occur mainly in lowland swamps, 75 (5.7 %) are shared with freshwater swamps and riparian habitats, 49 (3.7 %) are shared with heath forests, 7 (0.5 %) are shared with montane ecosystems, and 86 (6.5 %) are shared with a range of other lowland habitats. Of the 216 species (16.2 %) that occur in lowland swamps, 120 (9.2 %) are restricted to this habitat (which includes freshwater swamps), and 45 (3.4 %) are restricted to lowland peat swamp forests. Thus, more than 80 % (1,152 species) of the known peat swamp forest flora is common to a wide range of habitats, while 12.4 % (166 species) is -

Gone Fishing a Storytime About Fish
GONE FISHING A STORYTIME ABOUT FISH LESSON PLAN APRIL 2020 GONE FISHING A STORYTIME ABOUT FISH LESSON PLAN The objectives of this lesson are the following: ① to expand children’s curiosity and knowledge about fish, fishing and people who fish; ② to highlight the diversity of fish in the Great Lakes region and globally; ③ through reading, to connect children to the cultural and environmental attachments people have to fish and fishing; and ④ to introduce a few basic fish characteristics through a craft project. This lesson plan works well with children in preschool through second grade. The lesson lasts from 45 minutes to an hour, based on the number of books read. To orient the children to what it means to be a scientist, think scientifically and “do science,” use the “A Scientist Is...” and “Scientific Method” handouts available at the end of this lesson plan. Distribute fishing licenses found at the end of this lesson plan as the children arrive. GONE FISHING LESSON PLAN 1 SING Begin with your favorite welcome song. SCIENCE CHAT Begin by asking the children what they know about fish (e.g.,“Does anyone fish?” “What do you know about fish?” “Who eats fish?”). You can follow the brainstorm by sharing some of these facts about fish. FACTS ABOUT FISH: LOTS OF FISH, LOTS OF FACTS Fish are vertebrates (animals with a backbone, like humans). Fish make up about half of all the types (species) of backboned animals (vertebrates) existing today. Fish were the first vertebrates to live on earth and have existed for more than 500 million years. -

Evolutionary Trends of the Pharyngeal Dentition in Cypriniformes (Actinopterygii: Ostariophysi)
Evolutionary trends of the pharyngeal dentition in Cypriniformes (Actinopterygii: Ostariophysi). Emmanuel Pasco-Viel, Cyril Charles, Pascale Chevret, Marie Semon, Paul Tafforeau, Laurent Viriot, Vincent Laudet To cite this version: Emmanuel Pasco-Viel, Cyril Charles, Pascale Chevret, Marie Semon, Paul Tafforeau, et al.. Evolution- ary trends of the pharyngeal dentition in Cypriniformes (Actinopterygii: Ostariophysi).. PLoS ONE, Public Library of Science, 2010, 5 (6), pp.e11293. 10.1371/journal.pone.0011293. hal-00591939 HAL Id: hal-00591939 https://hal.archives-ouvertes.fr/hal-00591939 Submitted on 31 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Evolutionary Trends of the Pharyngeal Dentition in Cypriniformes (Actinopterygii: Ostariophysi) Emmanuel Pasco-Viel1, Cyril Charles3¤, Pascale Chevret2, Marie Semon2, Paul Tafforeau4, Laurent Viriot1,3*., Vincent Laudet2*. 1 Evo-devo of Vertebrate Dentition, Institut de Ge´nomique Fonctionnelle de Lyon, Universite´ de Lyon, CNRS, INRA, Ecole Normale Supe´rieure de Lyon, Lyon, France, 2 Molecular Zoology, Institut de Ge´nomique Fonctionnelle de Lyon, Universite´ de Lyon, CNRS, INRA, Ecole Normale Supe´rieure de Lyon, Lyon, France, 3 iPHEP, CNRS UMR 6046, Universite´ de Poitiers, Poitiers, France, 4 European Synchrotron Radiation Facility, Grenoble, France Abstract Background: The fish order Cypriniformes is one of the most diverse ray-finned fish groups in the world with more than 3000 recognized species. -

Emerging Semantics to Link Phenotype and Environment Anne E
Emerging semantics to link phenotype and environment Anne E. Thessen1,2 , Daniel E. Bunker3, Pier Luigi Buttigieg4, Laurel D. Cooper5, Wasila M. Dahdul6, Sami Domisch7, Nico M. Franz8, Pankaj Jaiswal5, Carolyn J. Lawrence-Dill9, Peter E. Midford10, Christopher J. Mungall11, Mart´ın J. Ram´ırez12, Chelsea D. Specht13, Lars Vogt14, Rutger Aldo Vos15, Ramona L. Walls16, JeVrey W. White17, Guanyang Zhang8, Andrew R. Deans18, Eva Huala19, Suzanna E. Lewis11 and Paula M. Mabee6 1 Ronin Institute for Independent Scholarship, Monclair, NJ, United States 2 The Data Detektiv, Waltham, MA, United States 3 Department of Biological Sciences, New Jersey Institute of Technology, Newark, NJ, United States 4 HGF-MPG Group for Deep Sea Ecology and Technology, Alfred-Wegener-Institut, Helmholtz-Zentrum fur¨ Polar-und Meeresforschung, Bremerhaven, Germany 5 Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR, United States 6 Department of Biology, University of South Dakota, Vermillion, SD, United States 7 Department of Ecology and Evolutionary Biology, Yale University, New Haven, CT, United States 8 School of Life Sciences, Arizona State University, Tempe, AZ, United States 9 Departments of Genetics, Development and Cell Biology and Agronomy, Iowa State University, Ames, IA, United States 10 Richmond, VA, United States 11 Genomics Division, Lawrence Berkeley National Laboratory, Berkeley, CA, United States 12 Division of Arachnology, Museo Argentino de Ciencias Naturales–CONICET, Buenos Aires, Argentina 13 Departments of Plant -

Resolving the Ray-Finned Fish Tree of Life COMMENTARY Michael E
COMMENTARY Resolving the ray-finned fish tree of life COMMENTARY Michael E. Alfaroa,1 The Challenge of Reconstructing Fish Phylogeny responsible for generating the trees in the first place When it comes to vertebrate evolutionary history, has emerged. In PNAS, Hughes et al. (12), use over our understanding of lobe-finned fishes—the branch 1,000 new markers using phylogenomic and tran- of the vertebrate tree leading to coelacanths plus scriptomic techniques to reconstruct the evolutionary the tetrapods (amphibians, turtles, birds, crocodiles, history of ray-finned fishes. Hughes et al.’s phylogeny lizards, snakes, and mammals)—far outstrips our reveals many key features of the backbone of the ray- knowledge of ray-finned fishes (Actinopterygii). Acti- finned fish tree of life and provides the most com- nopterygians exhibit extraordinary species richness prehensive evolutionary framework to date for this (>33,000 described species) and have evolved a stag- major radiation of vertebrates. gering diversity in morphology and ecology over their Phylogenomics has revolutionized evolutionary 400+ million y history. Ray-finned fishes include some reconstruction by vastly expanding both the number of the smallest vertebrates [the adult cyprinid Paedo- of loci that can be interrogated across nonmodel or- cypris progenetica measure just under 8 mm (1)], some ganisms, as well as the efficiency with which the data of the largest (adult ocean sunfish weigh more than may be collected (13). The most recent multilocus 2,000 kg), some of the longest (oarfishes may reach a studies that underlie our current understanding of the length of more than 13 m), some of the longest lived ray-finned fish tree of life are based upon 10– [rougheye rockfishes, Sebastes aleutianus, may live for 20 markers (e.g., refs. -

Ing.Org Published in Great Britain by the Royal Society, 6–9 Carlton House Terrace, London SW1Y 5AG Coturnix Coturnix Japonica Number 1665 Number
RSPB_276_1665_Cover.qxp 4/23/09 4:39 PM Page 1 Proc. R. Soc. B | vol. 276 no. 1665 pp. 2133–2331 22 June 2009 ISSN 0962-8452 volume 276 22 June 2009 number 1665 volume 276 . number 1665 . pages 2133–2331 pages 2133–2331 Review articles Chloroplast two-component systems: evolution of the link between photosynthesis and gene expression 2133 S. Puthiyaveetil & J. F. Allen Research articles Going to great lengths: selection for long corolla tubes in an extremely specialized bat–flower mutualism 2147 N. Muchhala & J. D. Thomson Dynamics of crowing development in the domestic Japanese quail (Coturnix coturnix japonica) 2153 S. Derégnaucourt, S. Saar & M. Gahr Emperor penguin mates: keeping together in the crowd 2163 A. Ancel, M. Beaulieu, Y. Le Maho & C. Gilbert A quantum probability explanation for violations of ‘rational’ decision theory 2171 E. M. Pothos & J. R. Busemeyer Spectacular morphological novelty in a miniature cyprinid fish, Danionella dracula n. sp. 2179 R. Britz, K. W. Conway & L. Rüber Does colour polymorphism enhance survival of prey populations? 2187 L. Wennersten & A. Forsman Genetic linkage map of the guppy, Poecilia reticulata, and quantitative trait loci analysis of male size and colour variation 2195 N. Tripathi, M. Hoffmann, E.-M. Willing, C. Lanz, D. Weigel & C. Dreyer Biodiversity and body size are linked across metazoans 2209 C. R. McClain & A. G. Boyer The evolution of covert, silent infection as a parasite strategy 2217 I. Sorrell, A. White, A. B. Pedersen, R. S. Hails & M. Boots Avian orientation: the pulse effect is mediated by the magnetite receptors in the upper beak 2227 W.