Activators and Effectors of the Small G Protein Arf1 in Regulation of Golgi Dynamics During the Cell Division Cycle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Novel Driver Strength Index Highlights Important Cancer Genes in TCGA Pancanatlas Patients
medRxiv preprint doi: https://doi.org/10.1101/2021.08.01.21261447; this version posted August 5, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . Novel Driver Strength Index highlights important cancer genes in TCGA PanCanAtlas patients Aleksey V. Belikov*, Danila V. Otnyukov, Alexey D. Vyatkin and Sergey V. Leonov Laboratory of Innovative Medicine, School of Biological and Medical Physics, Moscow Institute of Physics and Technology, 141701 Dolgoprudny, Moscow Region, Russia *Corresponding author: [email protected] NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. 1 medRxiv preprint doi: https://doi.org/10.1101/2021.08.01.21261447; this version posted August 5, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . Abstract Elucidating crucial driver genes is paramount for understanding the cancer origins and mechanisms of progression, as well as selecting targets for molecular therapy. Cancer genes are usually ranked by the frequency of mutation, which, however, does not necessarily reflect their driver strength. Here we hypothesize that driver strength is higher for genes that are preferentially mutated in patients with few driver mutations overall, because these few mutations should be strong enough to initiate cancer. -

Open Data for Differential Network Analysis in Glioma
International Journal of Molecular Sciences Article Open Data for Differential Network Analysis in Glioma , Claire Jean-Quartier * y , Fleur Jeanquartier y and Andreas Holzinger Holzinger Group HCI-KDD, Institute for Medical Informatics, Statistics and Documentation, Medical University Graz, Auenbruggerplatz 2/V, 8036 Graz, Austria; [email protected] (F.J.); [email protected] (A.H.) * Correspondence: [email protected] These authors contributed equally to this work. y Received: 27 October 2019; Accepted: 3 January 2020; Published: 15 January 2020 Abstract: The complexity of cancer diseases demands bioinformatic techniques and translational research based on big data and personalized medicine. Open data enables researchers to accelerate cancer studies, save resources and foster collaboration. Several tools and programming approaches are available for analyzing data, including annotation, clustering, comparison and extrapolation, merging, enrichment, functional association and statistics. We exploit openly available data via cancer gene expression analysis, we apply refinement as well as enrichment analysis via gene ontology and conclude with graph-based visualization of involved protein interaction networks as a basis for signaling. The different databases allowed for the construction of huge networks or specified ones consisting of high-confidence interactions only. Several genes associated to glioma were isolated via a network analysis from top hub nodes as well as from an outlier analysis. The latter approach highlights a mitogen-activated protein kinase next to a member of histondeacetylases and a protein phosphatase as genes uncommonly associated with glioma. Cluster analysis from top hub nodes lists several identified glioma-associated gene products to function within protein complexes, including epidermal growth factors as well as cell cycle proteins or RAS proto-oncogenes. -

Comprehensive Biological Information Analysis of PTEN Gene in Pan-Cancer
Comprehensive biological information analysis of PTEN gene in pan-cancer Hang Zhang Shanghai Medical University: Fudan University https://orcid.org/0000-0002-5853-7754 Wenhan Zhou Shanghai Medical University: Fudan University Xiaoyi Yang Shanghai Jiao Tong University School of Medicine Shuzhan Wen Shanghai Medical University: Fudan University Baicheng Zhao Shanghai Medical University: Fudan University Jiale Feng Shanghai Medical University: Fudan University Shuying Chen ( [email protected] ) https://orcid.org/0000-0002-9215-9777 Primary research Keywords: PTEN, correlated genes, TCGA, GEPIA, UALCAN, GTEx, expression, cancer Posted Date: April 12th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-388887/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/21 Abstract Background PTEN is a multifunctional tumor suppressor gene mutating at high frequency in a variety of cancers. However, its expression in pan-cancer, correlated genes, survival prognosis, and regulatory pathways are not completely described. Here, we aimed to conduct a comprehensive analysis from the above perspectives in order to provide reference for clinical application. Methods we studied the expression levels in cancers by using data from TCGA and GTEx database. Obtain expression box plot from UALCAN database. Perform mutation analysis on the cBioportal website. Obtain correlation genes on the GEPIA website. Construct protein network and perform KEGG and GO enrichment analysis on the STRING database. Perform prognostic analysis on the Kaplan-Meier Plotter website. We also performed transcription factor prediction on the PROMO database and performed RNA-RNA association and RNA-protein interaction on the RNAup Web server and RPISEq. -
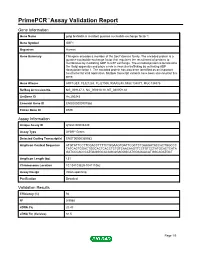
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name golgi brefeldin A resistant guanine nucleotide exchange factor 1 Gene Symbol GBF1 Organism Human Gene Summary This gene encodes a member of the Sec7 domain family. The encoded protein is a guanine nucleotide exchange factor that regulates the recruitment of proteins to membranes by mediating GDP to GTP exchange. The encoded protein is localized to the Golgi apparatus and plays a role in vesicular trafficking by activating ADP ribosylation factor 1. The encoded protein has also been identified as an important host factor for viral replication. Multiple transcript variants have been observed for this gene. Gene Aliases ARF1GEF, FLJ21263, FLJ21500, KIAA0248, MGC134877, MGC134878 RefSeq Accession No. NG_008147.1, NC_000010.10, NT_030059.13 UniGene ID Hs.290243 Ensembl Gene ID ENSG00000107862 Entrez Gene ID 8729 Assay Information Unique Assay ID qHsaCID0008440 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000369983 Amplicon Context Sequence ATGTATTCCTTCGACCTTTTCTGGAAGTGATTCGCTCTGAAGATACCACTGGCCC TATCACTGGACTGGCACTCACCTCTGTCAACAAGTTCCTGTCCTATGCACTCATA GATCCCACCCATGAGGGCACAGCAGAGGGCATGGAGAACATGGCAGATGCT Amplicon Length (bp) 131 Chromosome Location 10:104103829-104111062 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 94 R2 0.9988 cDNA Cq 20.43 cDNA Tm (Celsius) 84.5 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 36.2 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay -

Pharmacogenomics of Ventricular Conduction in Multi-Ethnic Populations Amanda Seyerle Dissertation Committee
Pharmacogenomics of Ventricular Conduction in Multi-Ethnic Populations Amanda Seyerle Dissertation Committee: Christy Avery (chair) Kari North Eric Whitsel Til Stürmer Craig Lee 1 TABLE OF CONTENTS Page LIST OF TABLES ...................................................................................................................................... v LIST OF FIGURES ..................................................................................................................................... vi LIST OF ABBREVIATIONS ..................................................................................................................... vii LIST OF GENE NAMES ............................................................................................................................ ix 1. Overview .............................................................................................................................................. 1 2. Specific Aims ....................................................................................................................................... 3 3. Background and Significance ............................................................................................................ 4 A. Ventricular Conduction ......................................................................................................................... 4 A.1. Electrical Conduction of the Heart ................................................................................................ 4 A.1.1. Sodium Channels.................................................................................................................. -

Gwas Meta-Analysis of Intelligence 1
bioRxiv preprint doi: https://doi.org/10.1101/184853; this version posted September 6, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. GWAS META-ANALYSIS OF INTELLIGENCE 1 GWAS meta-analysis (N=279,930) identifies new genes and functional links to intelligence Jeanne E Savage1#, Philip R Jansen1,2#, Sven Stringer1, Kyoko Watanabe1, Julien Bryois3, Christiaan A de Leeuw1, Mats Nagel1, Swapnil Awasthi4, Peter B Barr5, Jonathan R I Coleman6,7, Katrina L Grasby8, Anke R Hammerschlag1, Jakob Kaminski4,9, Robert Karlsson3, Eva Krapohl6, Max Lam10, Marianne Nygaard11,12, Chandra A Reynolds13, Joey W Trampush14, Hannah Young15, Delilah Zabaneh6, Sara Hägg3, Narelle K Hansell16, Ida K Karlsson3, Sten Linnarsson17, Grant W Montgomery8,18, Ana B Muñoz-Manchado17, Erin B Quinlan6, Gunter Schumann6, Nathan Skene17, Bradley T Webb19,20,21, Tonya White2, Dan E Arking22, Deborah K Attix23,24, Dimitrios Avramopoulos22,25, Robert M Bilder26, Panos Bitsios27, Katherine E Burdick28,29,30, Tyrone D Cannon31, Ornit Chiba-Falek32, Andrea Christoforou23, Elizabeth T Cirulli33, Eliza Congdon26, Aiden Corvin34, Gail Davies35, 36, Ian J Deary35,36, Pamela DeRosse37, Dwight Dickinson38, Srdjan Djurovic39,40, Gary Donohoe41, Emily Drabant Conley42, Johan G Eriksson43,44, Thomas Espeseth45,46, Nelson A Freimer26, Stella Giakoumaki47, Ina Giegling48, Michael Gill34, David -

Analyzing the Mirna-Gene Networks to Mine the Important Mirnas Under Skin of Human and Mouse
Hindawi Publishing Corporation BioMed Research International Volume 2016, Article ID 5469371, 9 pages http://dx.doi.org/10.1155/2016/5469371 Research Article Analyzing the miRNA-Gene Networks to Mine the Important miRNAs under Skin of Human and Mouse Jianghong Wu,1,2,3,4,5 Husile Gong,1,2 Yongsheng Bai,5,6 and Wenguang Zhang1 1 College of Animal Science, Inner Mongolia Agricultural University, Hohhot 010018, China 2Inner Mongolia Academy of Agricultural & Animal Husbandry Sciences, Hohhot 010031, China 3Inner Mongolia Prataculture Research Center, Chinese Academy of Science, Hohhot 010031, China 4State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650223, China 5Department of Biology, Indiana State University, Terre Haute, IN 47809, USA 6The Center for Genomic Advocacy, Indiana State University, Terre Haute, IN 47809, USA Correspondence should be addressed to Yongsheng Bai; [email protected] and Wenguang Zhang; [email protected] Received 11 April 2016; Revised 15 July 2016; Accepted 27 July 2016 Academic Editor: Nicola Cirillo Copyright © 2016 Jianghong Wu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Genetic networks provide new mechanistic insights into the diversity of species morphology. In this study, we have integrated the MGI, GEO, and miRNA database to analyze the genetic regulatory networks under morphology difference of integument of humans and mice. We found that the gene expression network in the skin is highly divergent between human and mouse. -

Rnai Screening Reveals Requirement for Host Cell Secretory Pathway in Infection by Diverse Families of Negative-Strand RNA Viruses
RNAi screening reveals requirement for host cell secretory pathway in infection by diverse families of negative-strand RNA viruses Debasis Pandaa,b, Anshuman Dasa,b, Phat X. Dinha,b, Sakthivel Subramaniama,b, Debasis Nayakc, Nicholas J. Barrowsd, James L. Pearsond, Jesse Thompsonb, David L. Kellye, Istvan Ladungaf, and Asit K. Pattnaika,b,1 aSchool of Veterinary Medicine and Biomedical Sciences and bNebraska Center for Virology, University of Nebraska, Lincoln, NE 68583; cNational Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892; dDuke RNAi Screening Facility, Duke University Medical Center, Durham, NC 27710; eUniversity of Nebraska Medical Center, Omaha, NE 68198; and fDepartment of Statistics, University of Nebraska, Lincoln, NE 68588 Edited by Peter Palese, Mount Sinai School of Medicine, New York, NY, and approved October 17, 2011 (received for review August 19, 2011) Negative-strand (NS) RNA viruses comprise many pathogens that and other NS RNA virus infections. Identifying the cellular factors cause serious diseases in humans and animals. Despite their clinical and studying the mechanisms of their involvement in these viral importance, little is known about the host factors required for their infections is important not only for understanding the biology of infection. Using vesicular stomatitis virus (VSV), a prototypic NS RNA these pathogens, but also for development of antiviral therapeutics. virus in the family Rhabdoviridae, we conducted a human genome- The advent of siRNA technology and the availability of ge- wide siRNA screen and identified 72 host genes required for viral nome-wide siRNA libraries have been useful in identifying host fl infection. Many of these identified genes were also required for factors required for in uenza virus, an NS RNA virus, and several – infection by two other NS RNA viruses, the lymphocytic choriome- positive-strand RNA viruses, as well as HIV (10 19). -

ARF4 Signalling Pathway Mediates the Response to Golgi Stress and Susceptibility to Pathogens
ARTICLES A CREB3–ARF4 signalling pathway mediates the response to Golgi stress and susceptibility to pathogens Jan H. Reiling1,2,3,6,7, Andrew J. Olive4, Sumana Sanyal1,2, Jan E. Carette1,6, Thijn R. Brummelkamp1,6, Hidde L. Ploegh1,2, Michael N. Starnbach4 and David M. Sabatini1,2,3,5,7 Treatment of cells with brefeldin A (BFA) blocks secretory vesicle transport and causes a collapse of the Golgi apparatus. To gain more insight into the cellular mechanisms mediating BFA toxicity, we conducted a genome-wide haploid genetic screen that led to the identification of the small G protein ADP-ribosylation factor 4 (ARF4). ARF4 depletion preserves viability, Golgi integrity and cargo trafficking in the presence of BFA, and these effects depend on the guanine nucleotide exchange factor GBF1 and other ARF isoforms including ARF1 and ARF5. ARF4 knockdown cells show increased resistance to several human pathogens including Chlamydia trachomatis and Shigella flexneri. Furthermore, ARF4 expression is induced when cells are exposed to several Golgi-disturbing agents and requires the CREB3 (also known as Luman or LZIP) transcription factor, whose downregulation mimics ARF4 loss. Thus, we have uncovered a CREB3–ARF4 signalling cascade that may be part of a Golgi stress response set in motion by stimuli compromising Golgi capacity. The processing, sorting and transport of proteins and lipids endocytic system by recruiting coat proteins, activating lipid-modifying in the Golgi to their destined intracellular site of action is of enzymes, and by regulating the cytoskeleton dynamics11,12. The fundamental importance to the maintenance and functions of effects of BFA are largely ascribed to the inhibition of ARF small G subcellular compartments. -

Genomic and Epigenomic Profile of Uterine Smooth Muscle
International Journal of Molecular Sciences Article Genomic and Epigenomic Profile of Uterine Smooth Muscle Tumors of Uncertain Malignant Potential (STUMPs) Revealed Similarities and Differences with Leiomyomas and Leiomyosarcomas Donatella Conconi 1,*,† , Serena Redaelli 1,† , Andrea Alberto Lissoni 1,2, Chiara Cilibrasi 3, Patrizia Perego 4, Eugenio Gautiero 5, Elena Sala 5, Mariachiara Paderno 1,2, Leda Dalprà 1, Fabio Landoni 1,2, Marialuisa Lavitrano 1 , Gaia Roversi 1,5 and Angela Bentivegna 1,* 1 School of Medicine and Surgery, University of Milano-Bicocca, 20900 Monza, Italy; [email protected] (S.R.); [email protected] (A.A.L.); [email protected] (M.P.); [email protected] (L.D.); [email protected] (F.L.); [email protected] (M.L.); [email protected] (G.R.) 2 Clinic of Obstetrics and Gynecology, San Gerardo Hospital, 20900 Monza, Italy 3 Department of Biochemistry and Biomedicine, School of Life Sciences, University of Sussex, Falmer, Brighton BN1 9RH, UK; [email protected] 4 Division of Pathology, San Gerardo Hospital, 20900 Monza, Italy; [email protected] 5 Medical Genetics Laboratory, San Gerardo Hospital, 20900 Monza, Italy; [email protected] (E.G.); [email protected] (E.S.) Citation: Conconi, D.; Redaelli, S.; * Correspondence: [email protected] (D.C.); [email protected] (A.B.); Lissoni, A.A.; Cilibrasi, C.; Perego, P.; Tel.: +39-0264488133 (A.B.) Gautiero, E.; Sala, E.; Paderno, M.; † Co-first authorship. Dalprà, L.; Landoni, F.; et al. Genomic and Epigenomic Profile of Uterine Abstract: Uterine smooth muscle tumors of uncertain malignant potential (STUMPs) represent a Smooth Muscle Tumors of Uncertain heterogeneous group of tumors that cannot be histologically diagnosed as unequivocally benign Malignant Potential (STUMPs) or malignant. -

Comparative Analysis of Eukaryotic Gene Sequence Features
Josep Francesc Abril Ferrando Comparative Analysis of Eukaryotic Gene Sequence Features Anàlisi Comparativa d’Elements de Seqüència dels Gens Eucariotes PhD Thesis Barcelona, May 2005 Dipòsit legal: B.47269-2005 ISBN: 978-84-691-1209-0 Comparative Analysis of Eukaryotic Gene Sequence Features Anàlisi Comparativa d’Elements de Seqüència dels Gens Eucariotes Josep Francesc Abril Ferrando PhD Thesis Barcelona, May 2005 CopyLeft 2005 by Josep Francesc Abril Ferrando. First Edition, April 2005. Printed at: COPISTERIA MIRACLE Rector Ubach, 6–10 (Aribau corner) 08021 — Barcelona Phone: +034 93 200 85 44 Fax: +034 93 209 17 82 Email: miracle at miraclepro.com Cover Figure: An artistic representation of how Bioinformatics helped to decode the human ge- nome. Metaphasic chromosomes are lying on top of a changing background where the DNA nucleic acids—A, C, G, and T, the language of life—, are con- verted into a binary code—0’s and 1’s, the language of computers—. A montage by J.F. Abril made with the Gimp (http://www.gimp.org/). Comparative Analysis of Eukaryotic Gene Sequence Features Anàlisi Comparativa d’Elements de Seqüència dels Gens Eucariotes Josep Francesc Abril Ferrando Memòria presentada per optar al grau de Doctor en Biologia per la Universitat Pompeu Fabra. Aquesta Tesi Doctoral ha estat realitzada sota la direcció del Dr. Roderic Guigó i Serra al Departament de Ciències Experimentals i de la Salut de la Universitat Pompeu Fabra. Roderic Guigó i Serra Josep Francesc Abril Ferrando Barcelona, May 2005 The research in this thesis has been carried out at the Genome BioInformatics Lab (GBIL) within the Grup de Recerca en Informàtica Biomèdica (GRIB) at the Parc de Re- cerca Biomèdica de Barcelona (PRBB), a consortium of the Institut Municipal d’Investigació Mèdica (IMIM), the Universitat Pompeu Fabra (UPF) and the Centre de Regulació Genòmica (CRG). -

Anti-PTGES2 (Aa 86-135) Polyclonal Antibody (DPABH-25479) This Product Is for Research Use Only and Is Not Intended for Diagnostic Use
Anti-PTGES2 (aa 86-135) polyclonal antibody (DPABH-25479) This product is for research use only and is not intended for diagnostic use. PRODUCT INFORMATION Antigen Description Isomerase that catalyzes the conversion of unstable intermediate of prostaglandin E2 H2 (PGH2) into the more stable prostaglandin E2 (PGE2) form. May also have transactivation activity toward IFN-gamma (IFNG), possibly via an interaction with CEBPB; however, the relevance of transcription activation activity remains unclear. Immunogen Synthetic peptide corresponding to a region within internal sequence amino acids 86-135 (VAKYMGAAAM YLISKRLKSR HRLQDNVRED LYEAADKWVA AVGKDRPFMG) of Human PTGES2 (NP_945176). Isotype IgG Source/Host Rabbit Species Reactivity Human Purification Immunogen affinity purified Conjugate Unconjugated Applications WB Format Liquid Size 50 μg Buffer Constituents: 97% PBS, 2% Sucrose Preservative None Storage Shipped at 4°C. Upon delivery aliquot and store at -20°C. Avoid repeated freeze / thaw cycles. GENE INFORMATION Gene Name PTGES2 prostaglandin E synthase 2 [ Homo sapiens ] Official Symbol PTGES2 Synonyms PTGES2; prostaglandin E synthase 2; C9orf15, chromosome 9 open reading frame 15; 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 1 © Creative Diagnostics All Rights Reserved FLJ14038; GATE-binding factor 1; microsomal prostaglandin E synthase-2; membrane- associated prostaglandin E synthase 2; gamma-interferon-activated transcriptional element- binding factor 1; GBF1;