Rnai Screening Reveals Requirement for Host Cell Secretory Pathway in Infection by Diverse Families of Negative-Strand RNA Viruses
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Transcriptome Response of High- and Low-Light-Adapted Prochlorococcus Strains to Changing Iron Availability
Transcriptome response of high- and low-light-adapted Prochlorococcus strains to changing iron availability The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Thompson, Anne W et al. “Transcriptome Response of High- and Low-light-adapted Prochlorococcus Strains to Changing Iron Availability.” ISME Journal (2011), 1-15. As Published http://dx.doi.org/10.1038/ismej.2011.49 Publisher Nature Publishing Group Version Author's final manuscript Citable link http://hdl.handle.net/1721.1/64705 Terms of Use Creative Commons Attribution-Noncommercial-Share Alike 3.0 Detailed Terms http://creativecommons.org/licenses/by-nc-sa/3.0/ Title: Transcriptome response of high- and low-light adapted Prochlorococcus strains to changing iron availability Running title: Prochlorococcus response to iron stress 5 Contributors: Anne W. Thompson1, Katherine Huang1, Mak A. Saito* 2, Sallie W. Chisholm* 1, 3 10 1 MIT Department of Civil and Environmental Engineering 2 Woods Hole Oceanographic Institution – Department of Marine Chemistry and Geochemistry 3 MIT Department of Biology 15 * To whom correspondence should be addressed: E-mail: [email protected] and [email protected] Subject Category: Microbial population and community ecology 20 Abstract Prochlorococcus contributes significantly to ocean primary productivity. The link between primary productivity and iron in specific ocean regions is well established and iron-limitation of Prochlorococcus cell division rates in these regions has been 25 demonstrated. However, the extent of ecotypic variation in iron metabolism among Prochlorococcus and the molecular basis for differences is not understood. Here, we examine the growth and transcriptional response of Prochlorococcus strains, MED4 and MIT9313, to changing iron concentrations. -

A Ph-Eqtl Interaction at the RIT2-SYT4 Parkinson's Disease Risk
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.16.423140; this version posted June 4, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. A pH-eQTL interaction at the RIT2-SYT4 Parkinson’s disease risk locus in the substantia nigra Authors and affiliations: Sejal Patel1*, Derek Howard1, Leon French1,2,3,4* 1. Krembil Centre for Neuroinformatics, Centre for Addiction and Mental Health, Toronto, ON, Canada 2. Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health, Toronto, Canada 3. Department of Psychiatry, University of Toronto, Toronto 4. Institute for Medical Science, University of Toronto, Toronto, Canada Abstract Parkinson's disease (PD) causes severe motor and cognitive disabilities that result from the progressive loss of dopamine neurons in the substantia nigra. The rs12456492 variant in the RIT2 gene has been repeatedly associated with increased risk for Parkinson's disease. From a transcriptomic perspective, a meta-analysis found that RIT2 gene expression is correlated with pH in the human brain. To assess these pH associations in relation to PD risk, we examined the two datasets that assayed rs12456492, gene expression, and pH in the postmortem human brain. Using the BrainEAC dataset, we replicate the positive correlation between RIT2 gene expression and pH in the human brain (n=100). Furthermore, we found that the relationship between expression and pH is influenced by rs12456492. -

View of HER2: Human Epidermal Growth Factor Receptor 2; TNBC: Triple-Negative Breast Resistance to Systemic Therapy in Patients with Breast Cancer
Wen et al. Cancer Cell Int (2018) 18:128 https://doi.org/10.1186/s12935-018-0625-9 Cancer Cell International PRIMARY RESEARCH Open Access Sulbactam‑enhanced cytotoxicity of doxorubicin in breast cancer cells Shao‑hsuan Wen1†, Shey‑chiang Su2†, Bo‑huang Liou3, Cheng‑hao Lin1 and Kuan‑rong Lee1* Abstract Background: Multidrug resistance (MDR) is a major obstacle in breast cancer treatment. The predominant mecha‑ nism underlying MDR is an increase in the activity of adenosine triphosphate (ATP)-dependent drug efux trans‑ porters. Sulbactam, a β-lactamase inhibitor, is generally combined with β-lactam antibiotics for treating bacterial infections. However, sulbactam alone can be used to treat Acinetobacter baumannii infections because it inhibits the expression of ATP-binding cassette (ABC) transporter proteins. This is the frst study to report the efects of sulbactam on mammalian cells. Methods: We used the breast cancer cell lines as a model system to determine whether sulbactam afects cancer cells. The cell viabilities in the present of doxorubicin with or without sulbactam were measured by MTT assay. Protein identities and the changes in protein expression levels in the cells after sulbactam and doxorubicin treatment were determined using LC–MS/MS. Real-time reverse transcription polymerase chain reaction (real-time RT-PCR) was used to analyze the change in mRNA expression levels of ABC transporters after treatment of doxorubicin with or without sulbactam. The efux of doxorubicin was measures by the doxorubicin efux assay. Results: MTT assay revealed that sulbactam enhanced the cytotoxicity of doxorubicin in breast cancer cells. The results of proteomics showed that ABC transporter proteins and proteins associated with the process of transcription and initiation of translation were reduced. -

Seq2pathway Vignette
seq2pathway Vignette Bin Wang, Xinan Holly Yang, Arjun Kinstlick May 19, 2021 Contents 1 Abstract 1 2 Package Installation 2 3 runseq2pathway 2 4 Two main functions 3 4.1 seq2gene . .3 4.1.1 seq2gene flowchart . .3 4.1.2 runseq2gene inputs/parameters . .5 4.1.3 runseq2gene outputs . .8 4.2 gene2pathway . 10 4.2.1 gene2pathway flowchart . 11 4.2.2 gene2pathway test inputs/parameters . 11 4.2.3 gene2pathway test outputs . 12 5 Examples 13 5.1 ChIP-seq data analysis . 13 5.1.1 Map ChIP-seq enriched peaks to genes using runseq2gene .................... 13 5.1.2 Discover enriched GO terms using gene2pathway_test with gene scores . 15 5.1.3 Discover enriched GO terms using Fisher's Exact test without gene scores . 17 5.1.4 Add description for genes . 20 5.2 RNA-seq data analysis . 20 6 R environment session 23 1 Abstract Seq2pathway is a novel computational tool to analyze functional gene-sets (including signaling pathways) using variable next-generation sequencing data[1]. Integral to this tool are the \seq2gene" and \gene2pathway" components in series that infer a quantitative pathway-level profile for each sample. The seq2gene function assigns phenotype-associated significance of genomic regions to gene-level scores, where the significance could be p-values of SNPs or point mutations, protein-binding affinity, or transcriptional expression level. The seq2gene function has the feasibility to assign non-exon regions to a range of neighboring genes besides the nearest one, thus facilitating the study of functional non-coding elements[2]. Then the gene2pathway summarizes gene-level measurements to pathway-level scores, comparing the quantity of significance for gene members within a pathway with those outside a pathway. -

Transcriptomic Analysis of the Aquaporin (AQP) Gene Family
Pancreatology 19 (2019) 436e442 Contents lists available at ScienceDirect Pancreatology journal homepage: www.elsevier.com/locate/pan Transcriptomic analysis of the Aquaporin (AQP) gene family interactome identifies a molecular panel of four prognostic markers in patients with pancreatic ductal adenocarcinoma Dimitrios E. Magouliotis a, b, Vasiliki S. Tasiopoulou c, Konstantinos Dimas d, * Nikos Sakellaridis d, Konstantina A. Svokos e, Alexis A. Svokos f, Dimitris Zacharoulis b, a Division of Surgery and Interventional Science, Faculty of Medical Sciences, UCL, London, UK b Department of Surgery, University of Thessaly, Biopolis, Larissa, Greece c Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece d Department of Pharmacology, Faculty of Medicine, School of Health Sciences, University of Thessaly, Biopolis, Larissa, Greece e The Warren Alpert Medical School of Brown University, Providence, RI, USA f Riverside Regional Medical Center, Newport News, VA, USA article info abstract Article history: Background: This study aimed to assess the differential gene expression of aquaporin (AQP) gene family Received 14 October 2018 interactome in pancreatic ductal adenocarcinoma (PDAC) using data mining techniques to identify novel Received in revised form candidate genes intervening in the pathogenicity of PDAC. 29 January 2019 Method: Transcriptome data mining techniques were used in order to construct the interactome of the Accepted 9 February 2019 AQP gene family and to determine which genes members are differentially expressed in PDAC as Available online 11 February 2019 compared to controls. The same techniques were used in order to evaluate the potential prognostic role of the differentially expressed genes. Keywords: PDAC Results: Transcriptome microarray data of four GEO datasets were incorporated, including 142 primary Aquaporin tumor samples and 104 normal pancreatic tissue samples. -

A Minimum-Labeling Approach for Reconstructing Protein Networks Across Multiple Conditions
A Minimum-Labeling Approach for Reconstructing Protein Networks across Multiple Conditions Arnon Mazza1, Irit Gat-Viks2, Hesso Farhan3, and Roded Sharan1 1 Blavatnik School of Computer Science, Tel Aviv University, Tel Aviv 69978, Israel. Email: [email protected]. 2 Dept. of Cell Research and Immunology, Tel Aviv University, Tel Aviv 69978, Israel. 3 Biotechnology Institute Thurgau, University of Konstanz, Unterseestrasse 47, CH-8280 Kreuzlingen, Switzerland. Abstract. The sheer amounts of biological data that are generated in recent years have driven the development of network analysis tools to fa- cilitate the interpretation and representation of these data. A fundamen- tal challenge in this domain is the reconstruction of a protein-protein sub- network that underlies a process of interest from a genome-wide screen of associated genes. Despite intense work in this area, current algorith- mic approaches are largely limited to analyzing a single screen and are, thus, unable to account for information on condition-specific genes, or reveal the dynamics (over time or condition) of the process in question. Here we propose a novel formulation for network reconstruction from multiple-condition data and devise an efficient integer program solution for it. We apply our algorithm to analyze the response to influenza in- fection in humans over time as well as to analyze a pair of ER export related screens in humans. By comparing to an extant, single-condition tool we demonstrate the power of our new approach in integrating data from multiple conditions in a compact and coherent manner, capturing the dynamics of the underlying processes. 1 Introduction With the increasing availability of high-throughput data, network biol- arXiv:1307.7803v1 [q-bio.QM] 30 Jul 2013 ogy has become the method of choice for filtering, interpreting and rep- resenting these data. -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -

Zbtb16 Regulates Social Cognitive Behaviors and Neocortical
Usui et al. Translational Psychiatry (2021) 11:242 https://doi.org/10.1038/s41398-021-01358-y Translational Psychiatry ARTICLE Open Access Zbtb16 regulates social cognitive behaviors and neocortical development Noriyoshi Usui 1,2,3,4, Stefano Berto5,AmiKonishi1, Makoto Kondo1,4, Genevieve Konopka5,HideoMatsuzaki 2,6,7 and Shoichi Shimada1,2,4 Abstract Zinc finger and BTB domain containing 16 (ZBTB16) play the roles in the neural progenitor cell proliferation and neuronal differentiation during development, however, how the function of ZBTB16 is involved in brain function and behaviors unknown. Here we show the deletion of Zbtb16 in mice leads to social impairment, repetitive behaviors, risk- taking behaviors, and cognitive impairment. To elucidate the mechanism underlying the behavioral phenotypes, we conducted histological analyses and observed impairments in thinning of neocortical layer 6 (L6) and a reduction of TBR1+ neurons in Zbtb16 KO mice. Furthermore, we found increased dendritic spines and microglia, as well as developmental defects in oligodendrocytes and neocortical myelination in the prefrontal cortex (PFC) of Zbtb16 KO mice. Using genomics approaches, we identified the Zbtb16 transcriptome that includes genes involved in neocortical maturation such as neurogenesis and myelination, and both autism spectrum disorder (ASD) and schizophrenia (SCZ) pathobiology. Co-expression networks further identified Zbtb16-correlated modules that are unique to ASD or SCZ, respectively. Our study provides insight into the novel roles of ZBTB16 in behaviors and neocortical development related to the disorders. 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; Introduction identified as a causative mutation for skeletal defects, ZBTB16 (PLZF) encodes a transcription factor, which genital hypoplasia, and mental retardation (SGYMR)6,7. -

Supplement 1 Microarray Studies
EASE Categories Significantly Enriched in vs MG vs vs MGC4-2 Pt1-C vs C4-2 Pt1-C UP-Regulated Genes MG System Gene Category EASE Global MGRWV Pt1-N RWV Pt1-N Score FDR GO Molecular Extracellular matrix cellular construction 0.0008 0 110 genes up- Function Interpro EGF-like domain 0.0009 0 regulated GO Molecular Oxidoreductase activity\ acting on single dono 0.0015 0 Function GO Molecular Calcium ion binding 0.0018 0 Function Interpro Laminin-G domain 0.0025 0 GO Biological Process Cell Adhesion 0.0045 0 Interpro Collagen Triple helix repeat 0.0047 0 KEGG pathway Complement and coagulation cascades 0.0053 0 KEGG pathway Immune System – Homo sapiens 0.0053 0 Interpro Fibrillar collagen C-terminal domain 0.0062 0 Interpro Calcium-binding EGF-like domain 0.0077 0 GO Molecular Cell adhesion molecule activity 0.0105 0 Function EASE Categories Significantly Enriched in Down-Regulated Genes System Gene Category EASE Global Score FDR GO Biological Process Copper ion homeostasis 2.5E-09 0 Interpro Metallothionein 6.1E-08 0 Interpro Vertebrate metallothionein, Family 1 6.1E-08 0 GO Biological Process Transition metal ion homeostasis 8.5E-08 0 GO Biological Process Heavy metal sensitivity/resistance 1.9E-07 0 GO Biological Process Di-, tri-valent inorganic cation homeostasis 6.3E-07 0 GO Biological Process Metal ion homeostasis 6.3E-07 0 GO Biological Process Cation homeostasis 2.1E-06 0 GO Biological Process Cell ion homeostasis 2.1E-06 0 GO Biological Process Ion homeostasis 2.1E-06 0 GO Molecular Helicase activity 2.3E-06 0 Function GO Biological -

An Arf1 Synthetic Lethal Screen Identifies a New Clathrin Heavy
Copyright 1998 by the Genetics Society of America An arf1D Synthetic Lethal Screen Identi®es a New Clathrin Heavy Chain Conditional Allele That Perturbs Vacuolar Protein Transport in Saccharomyces cerevisiae Chih-Ying Chen and Todd R. Graham Department of Molecular Biology, Vanderbilt University, Nashville, Tennessee 37235 Manuscript received March 5, 1998 Accepted for publication June 16, 1998 ABSTRACT ADP-ribosylation factor (ARF) is a small GTP-binding protein that is thought to regulate the assembly of coat proteins on transport vesicles. To identify factors that functionally interact with ARF, we have performed a genetic screen in Saccharomyces cerevisiae for mutations that exhibit synthetic lethality with an arf1D allele and de®ned seven genes by complementation tests (SWA1-7 for synthetically lethal with arf1D). Most of the swa mutants exhibit phenotypes comparable to arf1D mutants such as temperature-conditional growth, hypersensitivity to ¯uoride ions, and partial protein transport and glycosylation defects. Here, we report that swa5-1 is a new temperature-sensitive allele of the clathrin heavy chain gene (chc1-5), which carries a frameshift mutation near the 39 end of the CHC1 open reading frame. This genetic interaction between arf1 and chc1 provides in vivo evidence for a role for ARF in clathrin coat assembly. Surprisingly, strains harboring chc1-5 exhibited a signi®cant defect in transport of carboxypeptidase Y or carboxypepti- dase S to the vacuole that was not observed in other chc1 ts mutants. The kinetics of invertase secretion or transport of alkaline phosphatase to the vacuole were not signi®cantly affected in the chc1-5 mutant, further implicating clathrin speci®cally in the Golgi to vacuole transport pathway for carboxypeptidase Y. -
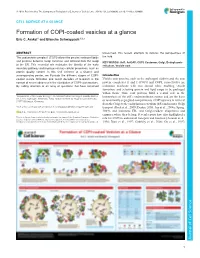
Formation of COPI-Coated Vesicles at a Glance Eric C
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs209890. doi:10.1242/jcs.209890 CELL SCIENCE AT A GLANCE Formation of COPI-coated vesicles at a glance Eric C. Arakel1 and Blanche Schwappach1,2,* ABSTRACT unresolved, this review attempts to refocus the perspectives of The coat protein complex I (COPI) allows the precise sorting of lipids the field. and proteins between Golgi cisternae and retrieval from the Golgi KEY WORDS: Arf1, ArfGAP, COPI, Coatomer, Golgi, Endoplasmic to the ER. This essential role maintains the identity of the early reticulum, Vesicle coat secretory pathway and impinges on key cellular processes, such as protein quality control. In this Cell Science at a Glance and accompanying poster, we illustrate the different stages of COPI- Introduction coated vesicle formation and revisit decades of research in the Vesicle coat proteins, such as the archetypal clathrin and the coat context of recent advances in the elucidation of COPI coat structure. protein complexes II and I (COPII and COPI, respectively) are By calling attention to an array of questions that have remained molecular machines with two central roles: enabling vesicle formation, and selecting protein and lipid cargo to be packaged within them. Thus, coat proteins fulfil a central role in the 1Department of Molecular Biology, Universitätsmedizin Göttingen, Humboldtallee homeostasis of the cell’s endomembrane system and are the basis 23, 37073 Göttingen, Germany. 2Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. of functionally segregated compartments. COPI operates in retrieval from the Golgi to the endoplasmic reticulum (ER) and in intra-Golgi *Author for correspondence ([email protected]) transport (Beck et al., 2009; Duden, 2003; Lee et al., 2004a; Spang, E.C.A., 0000-0001-7716-7149; B.S., 0000-0003-0225-6432 2009), and maintains ER- and Golgi-resident chaperones and enzymes where they belong. -

Anti-COPE Picoband Antibody Catalog # ABO13033
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Anti-COPE Picoband Antibody Catalog # ABO13033 Specification Anti-COPE Picoband Antibody - Product Information Application IHC Primary Accession O14579 Host Rabbit Reactivity Human, Mouse, Rat Clonality Polyclonal Format Lyophilized Description Rabbit IgG polyclonal antibody for Coatomer subunit epsilon(COPE) detection. Tested with WB, IHC-P in Human;Mouse;Rat. Figure 4. IHC analysis of COPE using Reconstitution anti-COPE antibody (ABO13033). Add 0.2ml of distilled water will yield a concentration of 500ug/ml. Anti-COPE Picoband Antibody - Additional Information Gene ID 11316 Other Names Coatomer subunit epsilon, Epsilon-coat protein, Epsilon-COP, COPE Calculated MW 34482 MW KDa Application Details Immunohistochemistry(Paraffin-embedded Section), 0.5-1 µg/ml, Human, Mouse, Rat, By Heat<br> Western blot, 0.1-0.5 µg/ml, Human, Mouse, Rat, <br> <br> Subcellular Localization Cytoplasm . Golgi apparatus membrane ; Peripheral membrane protein ; Cytoplasmic side . Cytoplasmic vesicle, COPI-coated vesicle membrane ; Peripheral membrane protein ; Cytoplasmic side . The coatomer is cytoplasmic or polymerized on the cytoplasmic side of the Golgi, as well as on the vesicles/buds originating from it. Page 1/3 10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 Contents Each vial contains 5mg BSA, 0.9mg NaCl, 0.2mg Na2HPO4, 0.05mg NaN3. Immunogen E. coli-derived human COPE recombinant protein (Position: E80-A308). Human COPE shares 89.5% amino acid (aa) sequence identity with mouse COPE. Purification Immunogen affinity purified. Cross Reactivity No cross reactivity with other proteins.