Analysis of Cdc6 Function in the Assembly of Mammalian Prereplication Complexes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Architecture of a Eukaryotic Replisome
The Architecture of a Eukaryotic Replisome Jingchuan Sun1,2, Yi Shi3, Roxana E. Georgescu3,4, Zuanning Yuan1,2, Brian T. Chait3, Huilin Li*1,2, Michael E. O’Donnell*3,4 1 Biosciences Department, Brookhaven National Laboratory, Upton, New York, USA 2 Department of Biochemistry & Cell Biology, Stony Brook University, Stony Brook, New York, USA. 3 The Rockefeller University, 1230 York Avenue, New York, New York, USA. 4 Howard Hughes Medical Institute *Correspondence and requests for materials should be addressed to M.O.D. ([email protected]) or H.L. ([email protected]) ABSTRACT At the eukaryotic DNA replication fork, it is widely believed that the Cdc45-Mcm2-7-GINS (CMG) helicase leads the way in front to unwind DNA, and that DNA polymerases (Pol) trail behind the helicase. Here we use single particle electron microscopy to directly image a replisome. Contrary to expectations, the leading strand Pol ε is positioned ahead of CMG helicase, while Ctf4 and the lagging strand Pol α-primase (Pol α) are behind the helicase. This unexpected architecture indicates that the leading strand DNA travels a long distance before reaching Pol ε, it first threads through the Mcm2-7 ring, then makes a U-turn at the bottom to reach Pol ε at the top of CMG. Our work reveals an unexpected configuration of the eukaryotic replisome, suggests possible reasons for this architecture, and provides a basis for further structural and biochemical replisome studies. INTRODUCTION DNA is replicated by a multi-protein machinery referred to as a replisome 1,2. Replisomes contain a helicase to unwind DNA, DNA polymerases that synthesize the leading and lagging strands, and a primase that makes short primed sites to initiate DNA synthesis on both strands. -

Orpl, a Member of the Cdcl8/Cdc6 Family of S-Phase Regulators, Is Homologous to a Component of the Origin Recognition Complex M
Proc. Natl. Acad. Sci. USA Vol. 92, pp. 12475-12479, December 1995 Genetics Orpl, a member of the Cdcl8/Cdc6 family of S-phase regulators, is homologous to a component of the origin recognition complex M. MuzI-FALCONI AND THOMAS J. KELLY Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine, Baltimore, MD 21205 Contributed by Thomas J. Kelly, September 11, 1995 ABSTRACT cdc18+ of Schizosaccharomyces pombe is a is the cdcJ8+ gene (1, 15). Expression of cdcJ8+ from a periodically expressed gene that is required for entry into S heterologous promoter is sufficient to rescue the lethality of a phase and for the coordination of S phase with mitosis. cdc18+ conditional temperature-sensitive (ts) cdc O's mutant. The is related to the Saccharomyces cerevisiae gene CDC6, which has cdcJ8+ gene product, a 65-kDa protein, is essential for the also been implicated in the control of DNA replication. We GI/S transition. Moreover, p65cdclS is a highly labile protein have identified a new Sch. pombe gene, orpl1, that encodes an whose expression is confined to a narrow window at the G,/S 80-kDa protein with amino acid sequence motifs conserved in boundary (unpublished data). These properties are consistent the Cdc18 and Cdc6 proteins. Genetic analysis indicates that with the hypothesis that Cdc18 may play an important role in orpi + is essential for viability. Germinating spores lacking the regulating the initiation of DNA replication at S phase. The orpl + gene are capable of undergoing one or more rounds of Cdc18 protein is homologous to the budding yeast Cdc6 DNA replication but fail to progress further, arresting as long protein, which may have a similar function (16). -

Derepression of Htert Gene Expression Promotes Escape from Oncogene-Induced Cellular Senescence
Derepression of hTERT gene expression promotes escape from oncogene-induced cellular senescence Priyanka L. Patela, Anitha Surama, Neena Miranib, Oliver Bischofc,d, and Utz Herbiga,e,1 aNew Jersey Medical School-Cancer Center, Rutgers Biomedical and Health Sciences, Newark, NJ 07103; bDepartment of Pathology and Laboratory Medicine, New Jersey Medical School, Rutgers Biomedical and Health Sciences, Newark, NJ 07103; cNuclear Organization and Oncogenesis Unit, Department of Cell Biology and Infection, Institut Pasteur, 75015 Paris, France; dINSERM U993, F-75015 Paris, France; and eDepartment of Microbiology, Biochemistry, and Molecular Genetics, Rutgers Biomedical and Health Sciences, Rutgers University, Newark, NJ 07103 Edited by Victoria Lundblad, Salk Institute for Biological Studies, La Jolla, CA, and approved June 27, 2016 (received for review February 11, 2016) Oncogene-induced senescence (OIS) is a critical tumor-suppressing that occur primarily at fragile sites. The ensuing DNA damage mechanism that restrains cancer progression at premalignant stages, response (DDR) triggers OIS, thereby arresting cells within a few in part by causing telomere dysfunction. Currently it is unknown cell-division cycles after oncogene expression (8, 9). Although most whether this proliferative arrest presents a stable and therefore DSBs in arrested cells are eventually resolved by cellular DSB irreversible barrier to cancer progression. Here we demonstrate that repair processes, some persist and consequently convert the other- cells frequently escape OIS induced by oncogenic H-Ras and B-Raf, wise transient DDR into a more permanent growth arrest. We and after a prolonged period in the senescence arrested state. Cells that others have demonstrated that the persistent DDR is primarily had escaped senescence displayed high oncogene expression levels, telomeric, triggered by irreparable telomeric DSBs (1, 10, 11). -

In Vivo Interactions of Archaeal Cdc6 Orc1 and Minichromosome
In vivo interactions of archaeal Cdc6͞Orc1 and minichromosome maintenance proteins with the replication origin Fujihiko Matsunaga*, Patrick Forterre*, Yoshizumi Ishino†, and Hannu Myllykallio*§ *Institut de Ge´ne´ tique et Microbiologie, Universite´de Paris-Sud, 91405 Orsay, France; and †Department of Molecular Biology, Biomolecular Engineering Research Institute, 6-2-3 Furuedai, Suita, Osaka 565-0874, Japan Communicated by Bruce W. Stillman, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, July 25, 2001 (received for review May 25, 2001) Although genome analyses have suggested parallels between basis of asymmetry in base composition between leading and archaeal and eukaryotic replication systems, little is known about lagging strands (10, 11). In Pyrococcus abyssi, the location of the the DNA replication mechanism in Archaea. By two-dimensional predicted origin coincides with an early replicating chromosomal gel electrophoreses we positioned a replication origin (oriC) within segment of 80 kb identified by radioactive labeling of chromo- 1 kb in the chromosomal DNA of Pyrococcus abyssi, an anaerobic somal DNA in cultures released from a replication block (12). hyperthermophile, and demonstrated that the oriC is physically Our in silico and labeling data also allowed us to conclude that linked to the cdc6 gene. Our chromatin immunoprecipitation as- the hyperthermophilic archaeon P. abyssi uses a single bidirec- says indicated that P. abyssi Cdc6 and minichromosome mainte- tional origin to replicate its genome. We proposed that this origin nance (MCM) proteins bind preferentially to the oriC region in the would correspond to the long intergenic region conserved in all exponentially growing cells. Whereas the oriC association of MCM three known Pyrococcus sp. -
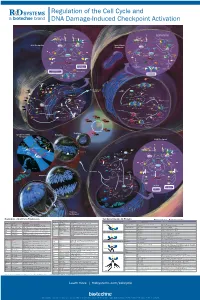
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

A Free-Living Protist That Lacks Canonical Eukaryotic DNA Replication and Segregation Systems
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.14.435266; this version posted March 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 A free-living protist that lacks canonical eukaryotic DNA replication and segregation systems 2 Dayana E. Salas-Leiva1, Eelco C. Tromer2,3, Bruce A. Curtis1, Jon Jerlström-Hultqvist1, Martin 3 Kolisko4, Zhenzhen Yi5, Joan S. Salas-Leiva6, Lucie Gallot-Lavallée1, Geert J. P. L. Kops3, John M. 4 Archibald1, Alastair G. B. Simpson7 and Andrew J. Roger1* 5 1Centre for Comparative Genomics and Evolutionary Bioinformatics (CGEB), Department of 6 Biochemistry and Molecular Biology, Dalhousie University, Halifax, NS, Canada, B3H 4R2 2 7 Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom 8 3Oncode Institute, Hubrecht Institute – KNAW (Royal Netherlands Academy of Arts and Sciences) 9 and University Medical Centre Utrecht, Utrecht, The Netherlands 10 4Institute of Parasitology Biology Centre, Czech Acad. Sci, České Budějovice, Czech Republic 11 5Guangzhou Key Laboratory of Subtropical Biodiversity and Biomonitoring, School of Life Science, 12 South China Normal University, Guangzhou 510631, China 13 6CONACyT-Centro de Investigación en Materiales Avanzados, Departamento de medio ambiente y 14 energía, Miguel de Cervantes 120, Complejo Industrial Chihuahua, 31136 Chihuahua, Chih., México 15 7Centre for Comparative Genomics and Evolutionary Bioinformatics (CGEB), Department of 16 Biology, Dalhousie University, Halifax, NS, Canada, B3H 4R2 17 *corresponding author: [email protected] 18 D.E.S-L ORCID iD: 0000-0003-2356-3351 19 E.C.T. -

Diversity of DNA Replication in the Archaea
G C A T T A C G G C A T genes Review Diversity of DNA Replication in the Archaea Darya Ausiannikava * and Thorsten Allers School of Life Sciences, University of Nottingham, Nottingham NG7 2UH, UK; [email protected] * Correspondence: [email protected]; Tel.: +44-115-823-0304 Academic Editor: Eishi Noguchi Received: 29 November 2016; Accepted: 20 January 2017; Published: 31 January 2017 Abstract: DNA replication is arguably the most fundamental biological process. On account of their shared evolutionary ancestry, the replication machinery found in archaea is similar to that found in eukaryotes. DNA replication is initiated at origins and is highly conserved in eukaryotes, but our limited understanding of archaea has uncovered a wide diversity of replication initiation mechanisms. Archaeal origins are sequence-based, as in bacteria, but are bound by initiator proteins that share homology with the eukaryotic origin recognition complex subunit Orc1 and helicase loader Cdc6). Unlike bacteria, archaea may have multiple origins per chromosome and multiple Orc1/Cdc6 initiator proteins. There is no consensus on how these archaeal origins are recognised—some are bound by a single Orc1/Cdc6 protein while others require a multi- Orc1/Cdc6 complex. Many archaeal genomes consist of multiple parts—the main chromosome plus several megaplasmids—and in polyploid species these parts are present in multiple copies. This poses a challenge to the regulation of DNA replication. However, one archaeal species (Haloferax volcanii) can survive without replication origins; instead, it uses homologous recombination as an alternative mechanism of initiation. This diversity in DNA replication initiation is all the more remarkable for having been discovered in only three groups of archaea where in vivo studies are possible. -

Scfcyclin F-Dependent Degradation of CDC6 Suppresses DNA Re-Replication
ARTICLE Received 3 Jun 2015 | Accepted 22 Dec 2015 | Published 28 Jan 2016 DOI: 10.1038/ncomms10530 OPEN SCFCyclin F-dependent degradation of CDC6 suppresses DNA re-replication David Walter1, Saskia Hoffmann1, Eirini-Stavroula Komseli2, Juri Rappsilber3,4, Vassilis Gorgoulis2,5 & Claus Storgaard Sørensen1 Maintenance of genome stability requires that DNA is replicated precisely once per cell cycle. This is believed to be achieved by limiting replication origin licensing and thereby restricting the firing of each replication origin to once per cell cycle. CDC6 is essential for eukaryotic replication origin licensing, however, it is poorly understood how CDC6 activity is constrained in higher eukaryotes. Here we report that the SCFCyclin F ubiquitin ligase complex prevents DNA re-replication by targeting CDC6 for proteasomal degradation late in the cell cycle. We show that CDC6 and Cyclin F interact through defined sequence motifs that promote CDC6 ubiquitylation and degradation. Absence of Cyclin F or expression of a stable mutant of CDC6 promotes re-replication and genome instability in cells lacking the CDT1 inhibitor Geminin. Together, our work reveals a novel SCFCyclin F-mediated mechanism required for precise once per cell cycle replication. 1 Biotech Research and Innovation Centre (BRIC), University of Copenhagen, Ole Maaløes Vej 5, Copenhagen N 2200, Denmark. 2 Department of Histology and Embryology, School of Medicine, University of Athens, Athens GR-11527, Greece. 3 Wellcome Trust Centre for Cell Biology, University of Edinburgh, Michael Swann Building, Kings Buildings, Max Born Crescent, Edinburgh EH9 3BF, Scotland. 4 Department of Bioanalytics, Institute of Biotechnology, Technische Universita¨t Berlin, Berlin 13355, Germany. -

PIP-Box-Mediated Degradation Prohibits Re-Accumulation of Cdc6 During S Phase
ß 2014. Published by The Company of Biologists Ltd | Journal of Cell Science (2014) 127, 1336–1345 doi:10.1242/jcs.145862 RESEARCH ARTICLE PIP-box-mediated degradation prohibits re-accumulation of Cdc6 during S phase Linda Clijsters1 and Rob Wolthuis1,2,* ABSTRACT levels of Cdt1 greatly increase owing to formation of a stable complex with its inhibitor geminin (Ballabeni et al., 2004; Cdc6 and Cdt1 initiate DNA replication licensing when cells exit Clijsters et al., 2013). When cells satisfy the spindle checkpoint mitosis. In cycling cells, Cdc6 is efficiently degraded from anaphase and exit mitosis, cyclin B1 is degraded through the E3 ligase onwards as a result of APC/C–Cdh1 activity. When APC/C–Cdh1 is anaphase-promoting complex/cyclosome containing Cdc20 switched off again, at the end of G1 phase, Cdc6 could thus re- (APC/C–Cdc20), which inactivates Cdk1, a key licensing accumulate, risking the re-licensing of DNA as long as Cdt1 is inhibitor (Pines, 2011). Geminin, a second key licensing present. Here, we carefully investigated the dynamics of Cdt1 and inhibitor in mammals, is also degraded by APC/C–Cdc20 when Cdc6 in cycling cells. We reveal a novel APC/C–Cdh1-independent the spindle checkpoint is satisfied (Clijsters et al., 2013). This degradation pathway that prevents nuclear Cdc6 re-accumulation at means that, in proliferating cells, the competence to license the G1-S transition and during S phase. Similar to Cdt1, nuclear DNA replication greatly increases at the metaphase-to-anaphase clearance of Cdc6 depends on an N-terminal PIP-box and the Cdt2- transition. -

The Architecture of the DNA Replication Origin Recognition Complex in Saccharomyces Cerevisiae
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Spiral - Imperial College Digital Repository The architecture of the DNA replication origin recognition complex in Saccharomyces cerevisiae Zhiqiang Chen, Christian Speck, Patricia Wendel, Chunyan Tang, Bruce Stillman, and Huilin Li ABSTRACT The origin recognition complex (ORC) is conserved in all eukaryotes. The six proteins of the Saccharomyces cerevisiae ORC that form a stable complex bind to origins of DNA replication and recruit prereplicative complex (pre-RC) proteins, one of which is Cdc6. To further understand the function of ORC we recently determined by single-particle reconstruction of electron micrographs a low-resolution, 3D structure of S. cerevisiae ORC and the ORC–Cdc6 complex. In this article, the spatial arrangement of the ORC subunits within the ORC structure is described. In one approach, a maltose binding protein (MBP) was systematically fused to the N or the C termini of the five largest ORC subunits, one subunit at a time, generating 10 MBP-fused ORCs, and the MBP density was localized in the averaged, 2D EM images of the MBP-fused ORC particles. Determining the Orc1–5 structure and comparing it with the native ORC structure localized the Orc6 subunit near Orc2 and Orc3. Finally, subunit–subunit interactions were determined by immunoprecipitation of ORC subunits synthesized in vitro. Based on the derived ORC architecture and existing structures of archaeal Orc1–DNA structures, we propose a model for ORC and suggest how ORC interacts with origin DNA and Cdc6. The studies provide a basis for understanding the overall structure of the pre- RC. -

Cdc6 on an Origin DNA Reveals the Mechanism of ORC
ARTICLE https://doi.org/10.1038/s41467-021-24199-1 OPEN The structure of ORC–Cdc6 on an origin DNA reveals the mechanism of ORC activation by the replication initiator Cdc6 ✉ ✉ Xiang Feng 1, Yasunori Noguchi2,3, Marta Barbon2,3, Bruce Stillman 4 , Christian Speck 2,3 & ✉ Huilin Li 1 1234567890():,; The Origin Recognition Complex (ORC) binds to sites in chromosomes to specify the location of origins of DNA replication. The S. cerevisiae ORC binds to specific DNA sequences throughout the cell cycle but becomes active only when it binds to the replication initiator Cdc6. It has been unclear at the molecular level how Cdc6 activates ORC, converting it to an active recruiter of the Mcm2-7 hexamer, the core of the replicative helicase. Here we report the cryo-EM structure at 3.3 Å resolution of the yeast ORC–Cdc6 bound to an 85-bp ARS1 origin DNA. The structure reveals that Cdc6 contributes to origin DNA recognition via its winged helix domain (WHD) and its initiator-specific motif. Cdc6 binding rearranges a short α-helix in the Orc1 AAA+ domain and the Orc2 WHD, leading to the activation of the Cdc6 ATPase and the formation of the three sites for the recruitment of Mcm2-7, none of which are present in ORC alone. The results illuminate the molecular mechanism of a critical biochemical step in the licensing of eukaryotic replication origins. 1 Department of Structural Biology, Van Andel Institute, Grand Rapids, MI, USA. 2 DNA Replication Group, Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, UK. -

Cdc6 Stability Is Regulated by the Huwe1 Ubiquitin Ligase After DNA Damage□D Jonathan R
Molecular Biology of the Cell Vol. 18, 3340–3350, September 2007 Cdc6 Stability Is Regulated by the Huwe1 Ubiquitin Ligase after DNA Damage□D Jonathan R. Hall,* Evelyn Kow,†‡ Kathleen R. Nevis,* Chiajung Karen Lu,§ K. Scott Luce,* Qing Zhong,§ and Jeanette Gowen Cook* *Department of Biochemistry and Biophysics, School of Medicine and Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC 27599-7260; †Department of Molecular Genetics, Duke University Medical Center, Durham, NC 27710; and §Division of Biochemistry and Molecular Biology, Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA 94720-3204 Submitted February 26, 2007; Revised May 9, 2007; Accepted June 6, 2007 Monitoring Editor: Orna Cohen-Fix The Cdc6 protein is an essential component of pre-replication complexes (preRCs), which assemble at origins of DNA replication during the G1 phase of the cell cycle. Previous studies have demonstrated that, in response to ionizing radiation, Cdc6 is ubiquitinated by the anaphase promoting complex (APCCdh1) in a p53-dependent manner. We find, however, that DNA damage caused by UV irradiation or DNA alkylation by methyl methane sulfonate (MMS) induces Cdc6 degradation independently of p53. We further demonstrate that Cdc6 degradation after these forms of DNA damage is also independent of cell cycle phase, Cdc6 phosphorylation of the known Cdk target residues, or the Cul4/DDB1 and APCCdh1 ubiquitin E3 ligases. Instead Cdc6 directly binds a HECT-family ubiquitin E3 ligase, Huwe1 (also known as Mule, UreB1, ARF-BP1, Lasu1, and HectH9), and Huwe1 polyubiquitinates Cdc6 in vitro. Degradation of Cdc6 in UV-irradiated cells or in cells treated with MMS requires Huwe1 and is associated with release of Cdc6 from chromatin.