The Architecture of the DNA Replication Origin Recognition Complex in Saccharomyces Cerevisiae
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

The Architecture of a Eukaryotic Replisome
The Architecture of a Eukaryotic Replisome Jingchuan Sun1,2, Yi Shi3, Roxana E. Georgescu3,4, Zuanning Yuan1,2, Brian T. Chait3, Huilin Li*1,2, Michael E. O’Donnell*3,4 1 Biosciences Department, Brookhaven National Laboratory, Upton, New York, USA 2 Department of Biochemistry & Cell Biology, Stony Brook University, Stony Brook, New York, USA. 3 The Rockefeller University, 1230 York Avenue, New York, New York, USA. 4 Howard Hughes Medical Institute *Correspondence and requests for materials should be addressed to M.O.D. ([email protected]) or H.L. ([email protected]) ABSTRACT At the eukaryotic DNA replication fork, it is widely believed that the Cdc45-Mcm2-7-GINS (CMG) helicase leads the way in front to unwind DNA, and that DNA polymerases (Pol) trail behind the helicase. Here we use single particle electron microscopy to directly image a replisome. Contrary to expectations, the leading strand Pol ε is positioned ahead of CMG helicase, while Ctf4 and the lagging strand Pol α-primase (Pol α) are behind the helicase. This unexpected architecture indicates that the leading strand DNA travels a long distance before reaching Pol ε, it first threads through the Mcm2-7 ring, then makes a U-turn at the bottom to reach Pol ε at the top of CMG. Our work reveals an unexpected configuration of the eukaryotic replisome, suggests possible reasons for this architecture, and provides a basis for further structural and biochemical replisome studies. INTRODUCTION DNA is replicated by a multi-protein machinery referred to as a replisome 1,2. Replisomes contain a helicase to unwind DNA, DNA polymerases that synthesize the leading and lagging strands, and a primase that makes short primed sites to initiate DNA synthesis on both strands. -

Orpl, a Member of the Cdcl8/Cdc6 Family of S-Phase Regulators, Is Homologous to a Component of the Origin Recognition Complex M
Proc. Natl. Acad. Sci. USA Vol. 92, pp. 12475-12479, December 1995 Genetics Orpl, a member of the Cdcl8/Cdc6 family of S-phase regulators, is homologous to a component of the origin recognition complex M. MuzI-FALCONI AND THOMAS J. KELLY Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine, Baltimore, MD 21205 Contributed by Thomas J. Kelly, September 11, 1995 ABSTRACT cdc18+ of Schizosaccharomyces pombe is a is the cdcJ8+ gene (1, 15). Expression of cdcJ8+ from a periodically expressed gene that is required for entry into S heterologous promoter is sufficient to rescue the lethality of a phase and for the coordination of S phase with mitosis. cdc18+ conditional temperature-sensitive (ts) cdc O's mutant. The is related to the Saccharomyces cerevisiae gene CDC6, which has cdcJ8+ gene product, a 65-kDa protein, is essential for the also been implicated in the control of DNA replication. We GI/S transition. Moreover, p65cdclS is a highly labile protein have identified a new Sch. pombe gene, orpl1, that encodes an whose expression is confined to a narrow window at the G,/S 80-kDa protein with amino acid sequence motifs conserved in boundary (unpublished data). These properties are consistent the Cdc18 and Cdc6 proteins. Genetic analysis indicates that with the hypothesis that Cdc18 may play an important role in orpi + is essential for viability. Germinating spores lacking the regulating the initiation of DNA replication at S phase. The orpl + gene are capable of undergoing one or more rounds of Cdc18 protein is homologous to the budding yeast Cdc6 DNA replication but fail to progress further, arresting as long protein, which may have a similar function (16). -

Derepression of Htert Gene Expression Promotes Escape from Oncogene-Induced Cellular Senescence
Derepression of hTERT gene expression promotes escape from oncogene-induced cellular senescence Priyanka L. Patela, Anitha Surama, Neena Miranib, Oliver Bischofc,d, and Utz Herbiga,e,1 aNew Jersey Medical School-Cancer Center, Rutgers Biomedical and Health Sciences, Newark, NJ 07103; bDepartment of Pathology and Laboratory Medicine, New Jersey Medical School, Rutgers Biomedical and Health Sciences, Newark, NJ 07103; cNuclear Organization and Oncogenesis Unit, Department of Cell Biology and Infection, Institut Pasteur, 75015 Paris, France; dINSERM U993, F-75015 Paris, France; and eDepartment of Microbiology, Biochemistry, and Molecular Genetics, Rutgers Biomedical and Health Sciences, Rutgers University, Newark, NJ 07103 Edited by Victoria Lundblad, Salk Institute for Biological Studies, La Jolla, CA, and approved June 27, 2016 (received for review February 11, 2016) Oncogene-induced senescence (OIS) is a critical tumor-suppressing that occur primarily at fragile sites. The ensuing DNA damage mechanism that restrains cancer progression at premalignant stages, response (DDR) triggers OIS, thereby arresting cells within a few in part by causing telomere dysfunction. Currently it is unknown cell-division cycles after oncogene expression (8, 9). Although most whether this proliferative arrest presents a stable and therefore DSBs in arrested cells are eventually resolved by cellular DSB irreversible barrier to cancer progression. Here we demonstrate that repair processes, some persist and consequently convert the other- cells frequently escape OIS induced by oncogenic H-Ras and B-Raf, wise transient DDR into a more permanent growth arrest. We and after a prolonged period in the senescence arrested state. Cells that others have demonstrated that the persistent DDR is primarily had escaped senescence displayed high oncogene expression levels, telomeric, triggered by irreparable telomeric DSBs (1, 10, 11). -

Supplementary Table S5. Differentially Expressed Gene Lists of PD-1High CD39+ CD8 Tils According to 4-1BB Expression Compared to PD-1+ CD39- CD8 Tils
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Immunother Cancer Supplementary Table S5. Differentially expressed gene lists of PD-1high CD39+ CD8 TILs according to 4-1BB expression compared to PD-1+ CD39- CD8 TILs Up- or down- regulated genes in Up- or down- regulated genes Up- or down- regulated genes only PD-1high CD39+ CD8 TILs only in 4-1BBneg PD-1high CD39+ in 4-1BBpos PD-1high CD39+ CD8 compared to PD-1+ CD39- CD8 CD8 TILs compared to PD-1+ TILs compared to PD-1+ CD39- TILs CD39- CD8 TILs CD8 TILs IL7R KLRG1 TNFSF4 ENTPD1 DHRS3 LEF1 ITGA5 MKI67 PZP KLF3 RYR2 SIK1B ANK3 LYST PPP1R3B ETV1 ADAM28 H2AC13 CCR7 GFOD1 RASGRP2 ITGAX MAST4 RAD51AP1 MYO1E CLCF1 NEBL S1PR5 VCL MPP7 MS4A6A PHLDB1 GFPT2 TNF RPL3 SPRY4 VCAM1 B4GALT5 TIPARP TNS3 PDCD1 POLQ AKAP5 IL6ST LY9 PLXND1 PLEKHA1 NEU1 DGKH SPRY2 PLEKHG3 IKZF4 MTX3 PARK7 ATP8B4 SYT11 PTGER4 SORL1 RAB11FIP5 BRCA1 MAP4K3 NCR1 CCR4 S1PR1 PDE8A IFIT2 EPHA4 ARHGEF12 PAICS PELI2 LAT2 GPRASP1 TTN RPLP0 IL4I1 AUTS2 RPS3 CDCA3 NHS LONRF2 CDC42EP3 SLCO3A1 RRM2 ADAMTSL4 INPP5F ARHGAP31 ESCO2 ADRB2 CSF1 WDHD1 GOLIM4 CDK5RAP1 CD69 GLUL HJURP SHC4 GNLY TTC9 HELLS DPP4 IL23A PITPNC1 TOX ARHGEF9 EXO1 SLC4A4 CKAP4 CARMIL3 NHSL2 DZIP3 GINS1 FUT8 UBASH3B CDCA5 PDE7B SOGA1 CDC45 NR3C2 TRIB1 KIF14 TRAF5 LIMS1 PPP1R2C TNFRSF9 KLRC2 POLA1 CD80 ATP10D CDCA8 SETD7 IER2 PATL2 CCDC141 CD84 HSPA6 CYB561 MPHOSPH9 CLSPN KLRC1 PTMS SCML4 ZBTB10 CCL3 CA5B PIP5K1B WNT9A CCNH GEM IL18RAP GGH SARDH B3GNT7 C13orf46 SBF2 IKZF3 ZMAT1 TCF7 NECTIN1 H3C7 FOS PAG1 HECA SLC4A10 SLC35G2 PER1 P2RY1 NFKBIA WDR76 PLAUR KDM1A H1-5 TSHZ2 FAM102B HMMR GPR132 CCRL2 PARP8 A2M ST8SIA1 NUF2 IL5RA RBPMS UBE2T USP53 EEF1A1 PLAC8 LGR6 TMEM123 NEK2 SNAP47 PTGIS SH2B3 P2RY8 S100PBP PLEKHA7 CLNK CRIM1 MGAT5 YBX3 TP53INP1 DTL CFH FEZ1 MYB FRMD4B TSPAN5 STIL ITGA2 GOLGA6L10 MYBL2 AHI1 CAND2 GZMB RBPJ PELI1 HSPA1B KCNK5 GOLGA6L9 TICRR TPRG1 UBE2C AURKA Leem G, et al. -

Congenital Microcephaly
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Sussex Research Online American Journal of Medical Genetics Part C (Seminars in Medical Genetics) ARTICLE Congenital Microcephaly DIANA ALCANTARA AND MARK O'DRISCOLL* The underlying etiologies of genetic congenital microcephaly are complex and multifactorial. Recently, with the exponential growth in the identification and characterization of novel genetic causes of congenital microcephaly, there has been a consolidation and emergence of certain themes concerning underlying pathomechanisms. These include abnormal mitotic microtubule spindle structure, numerical and structural abnormalities of the centrosome, altered cilia function, impaired DNA repair, DNA Damage Response signaling and DNA replication, along with attenuated cell cycle checkpoint proficiency. Many of these processes are highly interconnected. Interestingly, a defect in a gene whose encoded protein has a canonical function in one of these processes can often have multiple impacts at the cellular level involving several of these pathways. Here, we overview the key pathomechanistic themes underlying profound congenital microcephaly, and emphasize their interconnected nature. © 2014 Wiley Periodicals, Inc. KEY WORDS: cell division; mitosis; DNA replication; cilia How to cite this article: Alcantara D, O'Driscoll M. 2014. Congenital microcephaly. Am J Med Genet Part C Semin Med Genet 9999:1–16. INTRODUCTION mid‐gestation although glial cell division formation of the various cortical layers. and consequent brain volume enlarge- Furthermore, differentiating and devel- Congenital microcephaly, an occipital‐ ment does continue after birth [Spalding oping neurons must migrate to their frontal circumference of equal to or less et al., 2005]. Impaired neurogenesis is defined locations to construct the com- than 2–3 standard deviations below the therefore most obviously reflected clini- plex architecture and laminar layered age‐related population mean, denotes cally as congenital microcephaly. -

In Vivo Interactions of Archaeal Cdc6 Orc1 and Minichromosome
In vivo interactions of archaeal Cdc6͞Orc1 and minichromosome maintenance proteins with the replication origin Fujihiko Matsunaga*, Patrick Forterre*, Yoshizumi Ishino†, and Hannu Myllykallio*§ *Institut de Ge´ne´ tique et Microbiologie, Universite´de Paris-Sud, 91405 Orsay, France; and †Department of Molecular Biology, Biomolecular Engineering Research Institute, 6-2-3 Furuedai, Suita, Osaka 565-0874, Japan Communicated by Bruce W. Stillman, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, July 25, 2001 (received for review May 25, 2001) Although genome analyses have suggested parallels between basis of asymmetry in base composition between leading and archaeal and eukaryotic replication systems, little is known about lagging strands (10, 11). In Pyrococcus abyssi, the location of the the DNA replication mechanism in Archaea. By two-dimensional predicted origin coincides with an early replicating chromosomal gel electrophoreses we positioned a replication origin (oriC) within segment of 80 kb identified by radioactive labeling of chromo- 1 kb in the chromosomal DNA of Pyrococcus abyssi, an anaerobic somal DNA in cultures released from a replication block (12). hyperthermophile, and demonstrated that the oriC is physically Our in silico and labeling data also allowed us to conclude that linked to the cdc6 gene. Our chromatin immunoprecipitation as- the hyperthermophilic archaeon P. abyssi uses a single bidirec- says indicated that P. abyssi Cdc6 and minichromosome mainte- tional origin to replicate its genome. We proposed that this origin nance (MCM) proteins bind preferentially to the oriC region in the would correspond to the long intergenic region conserved in all exponentially growing cells. Whereas the oriC association of MCM three known Pyrococcus sp. -

Transcriptional Silencing Functions of the Yeast Protein Orc1/Sir3 Subfunctionalized After Gene Duplication
Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication Meleah A. Hickman1 and Laura N. Rusche2 Biochemistry Department, Institute for Genome Sciences and Policy, Program in Genetics and Genomics, Duke University, Durham, NC 27708 Edited by Jasper Rine, University of California, Berkeley, CA, and approved September 30, 2010 (received for review May 7, 2010) The origin recognition complex (ORC) defines origins of replication divergence is known (12, 13). We previously demonstrated that and also interacts with heterochromatin proteins in a variety of the duplicated deacetylases Sir2 and Hst1 subfunctionalized (14, species, but how ORC functions in heterochromatin assembly 15), and in this study, we propose that the SIR3–ORC1 gene pair remains unclear. The largest subunit of ORC, Orc1, is particularly subfunctionalized and then specialized after duplication. interesting because it contains a nucleosome-binding BAH domain In S. cerevisiae, SIR-mediated silencing occurs at telomeres and and because it gave rise to Sir3, a key silencing protein in Saccha- the mating-type loci, HMLα and HMRa (16). Transcriptional si- romyces cerevisiae, through gene duplication. We examined lencing of the mating-type loci is required to maintain cell-type whether Orc1 possessed a Sir3-like silencing function before du- identity in haploid cells. In contrast, SIR-silenced chromatin at plication and found that Orc1 from the yeast Kluyveromyces lactis, the telomeres is thought to serve a structural role (17). The Sir which diverged from S. cerevisiae before the duplication, acts in proteins are recruited to the mating-type loci through silencer conjunction with the deacetylase Sir2 and the histone-binding pro- sequences that bind ORC, Rap1, and Abf1, which in turn recruit tein Sir4 to generate heterochromatin at telomeres and a mating- the Sir proteins. -
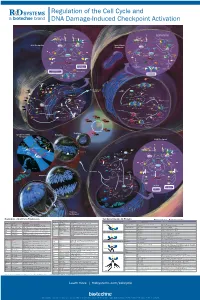
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

A Free-Living Protist That Lacks Canonical Eukaryotic DNA Replication and Segregation Systems
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.14.435266; this version posted March 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 A free-living protist that lacks canonical eukaryotic DNA replication and segregation systems 2 Dayana E. Salas-Leiva1, Eelco C. Tromer2,3, Bruce A. Curtis1, Jon Jerlström-Hultqvist1, Martin 3 Kolisko4, Zhenzhen Yi5, Joan S. Salas-Leiva6, Lucie Gallot-Lavallée1, Geert J. P. L. Kops3, John M. 4 Archibald1, Alastair G. B. Simpson7 and Andrew J. Roger1* 5 1Centre for Comparative Genomics and Evolutionary Bioinformatics (CGEB), Department of 6 Biochemistry and Molecular Biology, Dalhousie University, Halifax, NS, Canada, B3H 4R2 2 7 Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom 8 3Oncode Institute, Hubrecht Institute – KNAW (Royal Netherlands Academy of Arts and Sciences) 9 and University Medical Centre Utrecht, Utrecht, The Netherlands 10 4Institute of Parasitology Biology Centre, Czech Acad. Sci, České Budějovice, Czech Republic 11 5Guangzhou Key Laboratory of Subtropical Biodiversity and Biomonitoring, School of Life Science, 12 South China Normal University, Guangzhou 510631, China 13 6CONACyT-Centro de Investigación en Materiales Avanzados, Departamento de medio ambiente y 14 energía, Miguel de Cervantes 120, Complejo Industrial Chihuahua, 31136 Chihuahua, Chih., México 15 7Centre for Comparative Genomics and Evolutionary Bioinformatics (CGEB), Department of 16 Biology, Dalhousie University, Halifax, NS, Canada, B3H 4R2 17 *corresponding author: [email protected] 18 D.E.S-L ORCID iD: 0000-0003-2356-3351 19 E.C.T. -

Diversity of DNA Replication in the Archaea
G C A T T A C G G C A T genes Review Diversity of DNA Replication in the Archaea Darya Ausiannikava * and Thorsten Allers School of Life Sciences, University of Nottingham, Nottingham NG7 2UH, UK; [email protected] * Correspondence: [email protected]; Tel.: +44-115-823-0304 Academic Editor: Eishi Noguchi Received: 29 November 2016; Accepted: 20 January 2017; Published: 31 January 2017 Abstract: DNA replication is arguably the most fundamental biological process. On account of their shared evolutionary ancestry, the replication machinery found in archaea is similar to that found in eukaryotes. DNA replication is initiated at origins and is highly conserved in eukaryotes, but our limited understanding of archaea has uncovered a wide diversity of replication initiation mechanisms. Archaeal origins are sequence-based, as in bacteria, but are bound by initiator proteins that share homology with the eukaryotic origin recognition complex subunit Orc1 and helicase loader Cdc6). Unlike bacteria, archaea may have multiple origins per chromosome and multiple Orc1/Cdc6 initiator proteins. There is no consensus on how these archaeal origins are recognised—some are bound by a single Orc1/Cdc6 protein while others require a multi- Orc1/Cdc6 complex. Many archaeal genomes consist of multiple parts—the main chromosome plus several megaplasmids—and in polyploid species these parts are present in multiple copies. This poses a challenge to the regulation of DNA replication. However, one archaeal species (Haloferax volcanii) can survive without replication origins; instead, it uses homologous recombination as an alternative mechanism of initiation. This diversity in DNA replication initiation is all the more remarkable for having been discovered in only three groups of archaea where in vivo studies are possible. -

A Novel Motif Governs APC-Dependent Degradation of Drosophila ORC1 in Vivo
Downloaded from genesdev.cshlp.org on September 27, 2021 - Published by Cold Spring Harbor Laboratory Press A novel motif governs APC-dependent degradation of Drosophila ORC1 in vivo Marito Araki,1 Hongtao Yu,2 and Maki Asano1,3 1Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, North Carolina 27710, USA; 2Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, Texas 75390, USA Regulated degradation plays a key role in setting the level of many factors that govern cell cycle progression. In Drosophila, the largest subunit of the origin recognition complex protein 1 (ORC1) is degraded at the end of M phase and throughout much of G1 by anaphase-promoting complexes (APC) activated by Fzr/Cdh1. We show here that none of the previously identified APC motifs targets ORC1 for degradation. Instead, a novel sequence, the O-box, is necessary and sufficient to direct Fzr/Cdh1-dependent polyubiquitylation in vitro and degradation in vivo. The O-box is similar to but distinct from the well characterized D-box. Finally, we show that O-box motifs in two other proteins, Drosophila Abnormal Spindle and Schizosaccharomyces pombe Cut2, contribute to Cdh1-dependent polyubiquitylation in vitro, suggesting that the O-box may mediate degradation of a variety of cell cycle factors. [Keywords: APC; cell cycle; ORC1; proteolysis; replication] Received August 5, 2005; revised version accepted August 16, 2005. Progression through the cell cycle relies critically on the and Kirschner 2000), A-box (Littlepage and Ruderman scheduled degradation of an ensemble of proteins. Mis- 2002), and GxEN-box (Castro et al.