In Vivo Interactions of Archaeal Cdc6 Orc1 and Minichromosome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Thermodynamics of DNA Binding by DNA Polymerase I and Reca
Louisiana State University LSU Digital Commons LSU Doctoral Dissertations Graduate School 2014 Thermodynamics of DNA Binding by DNA Polymerase I and RecA Recombinase from Deinococcus radiodurans Jaycob Dalton Warfel Louisiana State University and Agricultural and Mechanical College Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_dissertations Recommended Citation Warfel, Jaycob Dalton, "Thermodynamics of DNA Binding by DNA Polymerase I and RecA Recombinase from Deinococcus radiodurans" (2014). LSU Doctoral Dissertations. 2382. https://digitalcommons.lsu.edu/gradschool_dissertations/2382 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Doctoral Dissertations by an authorized graduate school editor of LSU Digital Commons. For more information, please [email protected]. THERMODYNAMICS OF DNA BINDING BY DNA POLYMERASE I AND RECA RECOMBINASE FROM DEINOCOCCUS RADIODURANS A Dissertation Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Department of Biological Sciences by Jaycob Dalton Warfel B.S. Louisiana State University, 2006 May 2015 ACKNOWLEDGEMENTS I would like to express my utmost gratitude to the myriad of individuals who have lent their support during the time it has taken to complete this dissertation. First and foremost is due glory to God, The Father, The Son and The Holy Spirit, through whom all is accomplished. It is with extreme thankfulness for the blessings bestowed upon me, and with vast appreciation for the beauty of God’s creation that I have pursued a scientific education. -

The Architecture of a Eukaryotic Replisome
The Architecture of a Eukaryotic Replisome Jingchuan Sun1,2, Yi Shi3, Roxana E. Georgescu3,4, Zuanning Yuan1,2, Brian T. Chait3, Huilin Li*1,2, Michael E. O’Donnell*3,4 1 Biosciences Department, Brookhaven National Laboratory, Upton, New York, USA 2 Department of Biochemistry & Cell Biology, Stony Brook University, Stony Brook, New York, USA. 3 The Rockefeller University, 1230 York Avenue, New York, New York, USA. 4 Howard Hughes Medical Institute *Correspondence and requests for materials should be addressed to M.O.D. ([email protected]) or H.L. ([email protected]) ABSTRACT At the eukaryotic DNA replication fork, it is widely believed that the Cdc45-Mcm2-7-GINS (CMG) helicase leads the way in front to unwind DNA, and that DNA polymerases (Pol) trail behind the helicase. Here we use single particle electron microscopy to directly image a replisome. Contrary to expectations, the leading strand Pol ε is positioned ahead of CMG helicase, while Ctf4 and the lagging strand Pol α-primase (Pol α) are behind the helicase. This unexpected architecture indicates that the leading strand DNA travels a long distance before reaching Pol ε, it first threads through the Mcm2-7 ring, then makes a U-turn at the bottom to reach Pol ε at the top of CMG. Our work reveals an unexpected configuration of the eukaryotic replisome, suggests possible reasons for this architecture, and provides a basis for further structural and biochemical replisome studies. INTRODUCTION DNA is replicated by a multi-protein machinery referred to as a replisome 1,2. Replisomes contain a helicase to unwind DNA, DNA polymerases that synthesize the leading and lagging strands, and a primase that makes short primed sites to initiate DNA synthesis on both strands. -

Orpl, a Member of the Cdcl8/Cdc6 Family of S-Phase Regulators, Is Homologous to a Component of the Origin Recognition Complex M
Proc. Natl. Acad. Sci. USA Vol. 92, pp. 12475-12479, December 1995 Genetics Orpl, a member of the Cdcl8/Cdc6 family of S-phase regulators, is homologous to a component of the origin recognition complex M. MuzI-FALCONI AND THOMAS J. KELLY Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine, Baltimore, MD 21205 Contributed by Thomas J. Kelly, September 11, 1995 ABSTRACT cdc18+ of Schizosaccharomyces pombe is a is the cdcJ8+ gene (1, 15). Expression of cdcJ8+ from a periodically expressed gene that is required for entry into S heterologous promoter is sufficient to rescue the lethality of a phase and for the coordination of S phase with mitosis. cdc18+ conditional temperature-sensitive (ts) cdc O's mutant. The is related to the Saccharomyces cerevisiae gene CDC6, which has cdcJ8+ gene product, a 65-kDa protein, is essential for the also been implicated in the control of DNA replication. We GI/S transition. Moreover, p65cdclS is a highly labile protein have identified a new Sch. pombe gene, orpl1, that encodes an whose expression is confined to a narrow window at the G,/S 80-kDa protein with amino acid sequence motifs conserved in boundary (unpublished data). These properties are consistent the Cdc18 and Cdc6 proteins. Genetic analysis indicates that with the hypothesis that Cdc18 may play an important role in orpi + is essential for viability. Germinating spores lacking the regulating the initiation of DNA replication at S phase. The orpl + gene are capable of undergoing one or more rounds of Cdc18 protein is homologous to the budding yeast Cdc6 DNA replication but fail to progress further, arresting as long protein, which may have a similar function (16). -

Derepression of Htert Gene Expression Promotes Escape from Oncogene-Induced Cellular Senescence
Derepression of hTERT gene expression promotes escape from oncogene-induced cellular senescence Priyanka L. Patela, Anitha Surama, Neena Miranib, Oliver Bischofc,d, and Utz Herbiga,e,1 aNew Jersey Medical School-Cancer Center, Rutgers Biomedical and Health Sciences, Newark, NJ 07103; bDepartment of Pathology and Laboratory Medicine, New Jersey Medical School, Rutgers Biomedical and Health Sciences, Newark, NJ 07103; cNuclear Organization and Oncogenesis Unit, Department of Cell Biology and Infection, Institut Pasteur, 75015 Paris, France; dINSERM U993, F-75015 Paris, France; and eDepartment of Microbiology, Biochemistry, and Molecular Genetics, Rutgers Biomedical and Health Sciences, Rutgers University, Newark, NJ 07103 Edited by Victoria Lundblad, Salk Institute for Biological Studies, La Jolla, CA, and approved June 27, 2016 (received for review February 11, 2016) Oncogene-induced senescence (OIS) is a critical tumor-suppressing that occur primarily at fragile sites. The ensuing DNA damage mechanism that restrains cancer progression at premalignant stages, response (DDR) triggers OIS, thereby arresting cells within a few in part by causing telomere dysfunction. Currently it is unknown cell-division cycles after oncogene expression (8, 9). Although most whether this proliferative arrest presents a stable and therefore DSBs in arrested cells are eventually resolved by cellular DSB irreversible barrier to cancer progression. Here we demonstrate that repair processes, some persist and consequently convert the other- cells frequently escape OIS induced by oncogenic H-Ras and B-Raf, wise transient DDR into a more permanent growth arrest. We and after a prolonged period in the senescence arrested state. Cells that others have demonstrated that the persistent DDR is primarily had escaped senescence displayed high oncogene expression levels, telomeric, triggered by irreparable telomeric DSBs (1, 10, 11). -

Access to Electronic Thesis
Access to Electronic Thesis Author: Khalid Salim Al-Abri Thesis title: USE OF MOLECULAR APPROACHES TO STUDY THE OCCURRENCE OF EXTREMOPHILES AND EXTREMODURES IN NON-EXTREME ENVIRONMENTS Qualification: PhD This electronic thesis is protected by the Copyright, Designs and Patents Act 1988. No reproduction is permitted without consent of the author. It is also protected by the Creative Commons Licence allowing Attributions-Non-commercial-No derivatives. If this electronic thesis has been edited by the author it will be indicated as such on the title page and in the text. USE OF MOLECULAR APPROACHES TO STUDY THE OCCURRENCE OF EXTREMOPHILES AND EXTREMODURES IN NON-EXTREME ENVIRONMENTS By Khalid Salim Al-Abri Msc., University of Sultan Qaboos, Muscat, Oman Mphil, University of Sheffield, England Thesis submitted in partial fulfillment for the requirements of the Degree of Doctor of Philosophy in the Department of Molecular Biology and Biotechnology, University of Sheffield, England 2011 Introductory Pages I DEDICATION To the memory of my father, loving mother, wife “Muneera” and son “Anas”, brothers and sisters. Introductory Pages II ACKNOWLEDGEMENTS Above all, I thank Allah for helping me in completing this project. I wish to express my thanks to my supervisor Professor Milton Wainwright, for his guidance, supervision, support, understanding and help in this project. In addition, he also stood beside me in all difficulties that faced me during study. My thanks are due to Dr. D. J. Gilmour for his co-supervision, technical assistance, his time and understanding that made some of my laboratory work easier. In the Ministry of Regional Municipalities and Water Resources, I am particularly grateful to Engineer Said Al Alawi, Director General of Health Control, for allowing me to carry out my PhD study at the University of Sheffield. -

Representatives of a Novel Archaeal Phylum Or a Fast-Evolving
Open Access Research2005BrochieretVolume al. 6, Issue 5, Article R42 Nanoarchaea: representatives of a novel archaeal phylum or a comment fast-evolving euryarchaeal lineage related to Thermococcales? Celine Brochier*, Simonetta Gribaldo†, Yvan Zivanovic‡, Fabrice Confalonieri‡ and Patrick Forterre†‡ Addresses: *EA EGEE (Evolution, Génomique, Environnement) Université Aix-Marseille I, Centre Saint-Charles, 3 Place Victor Hugo, 13331 Marseille, Cedex 3, France. †Unite Biologie Moléculaire du Gène chez les Extremophiles, Institut Pasteur, 25 rue du Dr Roux, 75724 Paris Cedex ‡ 15, France. Institut de Génétique et Microbiologie, UMR CNRS 8621, Université Paris-Sud, 91405 Orsay, France. reviews Correspondence: Celine Brochier. E-mail: [email protected]. Simonetta Gribaldo. E-mail: [email protected] Published: 14 April 2005 Received: 3 December 2004 Revised: 10 February 2005 Genome Biology 2005, 6:R42 (doi:10.1186/gb-2005-6-5-r42) Accepted: 9 March 2005 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2005/6/5/R42 reports © 2005 Brochier et al.; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Placement<p>Anteins from analysis 25of Nanoarcheumarchaeal of the positiongenomes equitans of suggests Nanoarcheum in the that archaeal N. equitans phylogeny inis likethe lyarchaeal to be the phylogeny representative using aof large a fast-evolving dataset of concatenatedeuryarchaeal ribosomalineage.</p>l pro- deposited research Abstract Background: Cultivable archaeal species are assigned to two phyla - the Crenarchaeota and the Euryarchaeota - by a number of important genetic differences, and this ancient split is strongly supported by phylogenetic analysis. -

Tackling the Methanopyrus Kandleri Paradox Céline Brochier*, Patrick Forterre† and Simonetta Gribaldo†
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central Open Access Research2004BrochieretVolume al. 5, Issue 3, Article R17 Archaeal phylogeny based on proteins of the transcription and comment translation machineries: tackling the Methanopyrus kandleri paradox Céline Brochier*, Patrick Forterre† and Simonetta Gribaldo† Addresses: *Equipe Phylogénomique, Université Aix-Marseille I, Centre Saint-Charles, 13331 Marseille Cedex 3, France. †Institut de Génétique et Microbiologie, CNRS UMR 8621, Université Paris-Sud, 91405 Orsay, France. reviews Correspondence: Céline Brochier. E-mail: [email protected] Published: 26 February 2004 Received: 14 November 2003 Revised: 5 January 2004 Genome Biology 2004, 5:R17 Accepted: 21 January 2004 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2004/5/3/R17 reports © 2004 Brochier et al.; licensee BioMed Central Ltd. This is an Open Access article: verbatim copying and redistribution of this article are permitted in all media for any purpose, provided this notice is preserved along with the article's original URL. ArchaealPhylogeneticsequencedusingrespectively). two phylogeny concatenated genomes, analysis based it of is datasetsthe now on Archaea proteinspossible consisting has ofto been thetest of transcription alternative mainly14 proteins established approach involv and translationed byes in 16S bytranscription rRNAusing machineries: largesequence andsequence 53comparison.tackling ribosomal datasets. the Methanopyrus Withproteins We theanalyzed accumulation(3,275 archaealkandleri and 6,377 of phyparadox comp positions,logenyletely Abstract deposited research Background: Phylogenetic analysis of the Archaea has been mainly established by 16S rRNA sequence comparison. With the accumulation of completely sequenced genomes, it is now possible to test alternative approaches by using large sequence datasets. -

Microorganisms
microorganisms Article Comparative Proteomics Analysis Reveals New Features of the Oxidative Stress Response in the Polyextremophilic Bacterium Deinococcus radiodurans Lihua Gao, Zhengfu Zhou, Xiaonan Chen, Wei Zhang, Min Lin and Ming Chen * Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, China; [email protected] (L.G.); [email protected] (Z.Z.); [email protected] (X.C.); [email protected] (W.Z.); [email protected] (M.L.) * Correspondence: [email protected] Received: 20 February 2020; Accepted: 19 March 2020; Published: 23 March 2020 Abstract: Deinococcus radiodurans is known for its extreme resistance to ionizing radiation, oxidative stress, and other DNA-damaging agents. The robustness of this bacterium primarily originates from its strong oxidative resistance mechanisms. Hundreds of genes have been demonstrated to contribute to oxidative resistance in D. radiodurans; however, the antioxidant mechanisms have not been fully characterized. In this study, comparative proteomics analysis of D. radiodurans grown under normal and oxidative stress conditions was conducted using label-free quantitative proteomics. The abundances of 852 of 1700 proteins were found to significantly differ between the two groups. These differential proteins are mainly associated with translation, DNA repair and recombination, response to stresses, transcription, and cell wall organization. Highly upregulated expression was observed for ribosomal proteins such as RplB, Rpsl, RpsR, DNA damage response proteins (DdrA, DdrB), DNA repair proteins (RecN, RecA), and transcriptional regulators (members of TetR, AsnC, and GntR families, DdrI). The functional analysis of proteins in response to oxidative stress is discussed in detail. This study reveals the global protein expression profile of D. radiodurans in response to oxidative stress and provides new insights into the regulatory mechanism of oxidative resistance in D. -
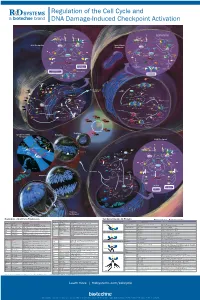
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

A Free-Living Protist That Lacks Canonical Eukaryotic DNA Replication and Segregation Systems
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.14.435266; this version posted March 15, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 A free-living protist that lacks canonical eukaryotic DNA replication and segregation systems 2 Dayana E. Salas-Leiva1, Eelco C. Tromer2,3, Bruce A. Curtis1, Jon Jerlström-Hultqvist1, Martin 3 Kolisko4, Zhenzhen Yi5, Joan S. Salas-Leiva6, Lucie Gallot-Lavallée1, Geert J. P. L. Kops3, John M. 4 Archibald1, Alastair G. B. Simpson7 and Andrew J. Roger1* 5 1Centre for Comparative Genomics and Evolutionary Bioinformatics (CGEB), Department of 6 Biochemistry and Molecular Biology, Dalhousie University, Halifax, NS, Canada, B3H 4R2 2 7 Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom 8 3Oncode Institute, Hubrecht Institute – KNAW (Royal Netherlands Academy of Arts and Sciences) 9 and University Medical Centre Utrecht, Utrecht, The Netherlands 10 4Institute of Parasitology Biology Centre, Czech Acad. Sci, České Budějovice, Czech Republic 11 5Guangzhou Key Laboratory of Subtropical Biodiversity and Biomonitoring, School of Life Science, 12 South China Normal University, Guangzhou 510631, China 13 6CONACyT-Centro de Investigación en Materiales Avanzados, Departamento de medio ambiente y 14 energía, Miguel de Cervantes 120, Complejo Industrial Chihuahua, 31136 Chihuahua, Chih., México 15 7Centre for Comparative Genomics and Evolutionary Bioinformatics (CGEB), Department of 16 Biology, Dalhousie University, Halifax, NS, Canada, B3H 4R2 17 *corresponding author: [email protected] 18 D.E.S-L ORCID iD: 0000-0003-2356-3351 19 E.C.T. -

Druschel Et Al EG V.97.Pdf
Economic Geology Vol. 97, 2002, pp. 0000–0000 Geochemical Modeling of ZnS in Biofilms: An Example of Ore Depositional Processes G. K. DRUSCHEL,† M. LABRENZ,* T. THOMSEN-EBERT, D. A. FOWLE,** AND J. F. BANFIELD*** Department of Geology and Geophysics, University of Wisconsin-Madison, 1215 W. Dayton St., Madison, WI 53706 Abstract The precipitation of nearly pure, nanocrystalline zinc sulfides (primarily sphalerite and wurtzite) within a biofilm dominated by sulfate-reducing bacteria of the family Desulfobacteriaceae has been observed in the flooded tunnels of an abandoned mine in southwestern Wisconsin. ZnS accumulations are limited to biofilms growing on old mine timbers. ZnS deposits, which comprise about 20 percent of the volume of the biofilm, formed in less than 30 yr from solutions containing only a few milligrams per liter (mg/l) Zn2+. A model is pro- posed wherein sulfate reduction is followed by the formation of aqueous metal-sulfide clusters that aggregate to form 1- to 3-nm–diameter crystals that are transported and agglomerate to form micron-scale aggre- gates. Geochemical modeling shows how reduction of sulfate leads to exclusive precipitation of ZnS until most of the Zn2+ is consumed. The model also predicts a series of discrete metal-sulfide precipitation events that may be used to interpret sulfide mineral paragenesis of low-temperature Cu-Pb-Zn ore deposits. For example, the paragenesis of essentially monomineralic and mixed sulfide layers in Mississippi Valley-type, strataform, and strata-bound Pb-Zn-Cu, and SEDEX deposits can be predicted thermodynamically if the rate of aqueous sul- fide generation does not outstrip the flux of metals into the system. -

Diversity of DNA Replication in the Archaea
G C A T T A C G G C A T genes Review Diversity of DNA Replication in the Archaea Darya Ausiannikava * and Thorsten Allers School of Life Sciences, University of Nottingham, Nottingham NG7 2UH, UK; [email protected] * Correspondence: [email protected]; Tel.: +44-115-823-0304 Academic Editor: Eishi Noguchi Received: 29 November 2016; Accepted: 20 January 2017; Published: 31 January 2017 Abstract: DNA replication is arguably the most fundamental biological process. On account of their shared evolutionary ancestry, the replication machinery found in archaea is similar to that found in eukaryotes. DNA replication is initiated at origins and is highly conserved in eukaryotes, but our limited understanding of archaea has uncovered a wide diversity of replication initiation mechanisms. Archaeal origins are sequence-based, as in bacteria, but are bound by initiator proteins that share homology with the eukaryotic origin recognition complex subunit Orc1 and helicase loader Cdc6). Unlike bacteria, archaea may have multiple origins per chromosome and multiple Orc1/Cdc6 initiator proteins. There is no consensus on how these archaeal origins are recognised—some are bound by a single Orc1/Cdc6 protein while others require a multi- Orc1/Cdc6 complex. Many archaeal genomes consist of multiple parts—the main chromosome plus several megaplasmids—and in polyploid species these parts are present in multiple copies. This poses a challenge to the regulation of DNA replication. However, one archaeal species (Haloferax volcanii) can survive without replication origins; instead, it uses homologous recombination as an alternative mechanism of initiation. This diversity in DNA replication initiation is all the more remarkable for having been discovered in only three groups of archaea where in vivo studies are possible.