Improved Forensic Toxicology Screening Using a GC/MS/NPD System with a 725-Compound DRS Database
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Basic Quant GCMS
Harris County Institute of Forensic Sciences Section: Toxicology Approved By: Toxicology Manager Document Type: GC & GC/MS Procedure No.: TOX07.3005 Title: Quantitation of basic drugs by gas chromatography / mass spectrometry Rev.:15 1.0 Purpose 1.1 This document describes the procedures used by the Toxicology Laboratory to quantitate basic drugs using gas chromatography/mass spectrometry. 1.2 Biological specimens are basified to the moderately alkaline pH 9 at which basic compounds are in an unionized form and are soluble in an organic solvent. After liquid/liquid extraction, the organic layer is separated and the compound X is back extracted into 2 N HCl. The acid layer is then made alkaline and compound X is re-extracted into an organic solvent. 2.0 Scope 2.1 The assay is appropriate for quantitation of any basic compound (X) in blood including serum and plasma, urine, bile, stomach contents or tissue homogenates. 3.0 Definitions and Abbreviations 3.1 No method-specific or non-standard terms are used in this procedure. 4.0 Materials 4.1 Instruments and Equipment 4.1.1 13 x 100 mm screw top tubes and caps 4.1.2 Mixer 4.1.3 Rocker 4.1.4 Centrifuge 4.1.5 Transfer pipettes 4.1.6 Autosampler vials and rubber septum caps 4.1.7 Autosampler vial inserts 4.1.8 Vial crimper 4.1.9 100uL syringe 4.1.10 GC/MS Uncontrolled4.2 Reagents Copy 4.2.1 Ammonium Hydroxide Reagent Grade 4.2.2 n-Butyl chloride (chlorobutane) HPLC Grade 4.2.3 Saturated Sodium Borate Buffer, pH 9.3 A. -

(12) Patent Application Publication (10) Pub. No.: US 2006/0024365A1 Vaya Et Al
US 2006.0024.365A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2006/0024365A1 Vaya et al. (43) Pub. Date: Feb. 2, 2006 (54) NOVEL DOSAGE FORM (30) Foreign Application Priority Data (76) Inventors: Navin Vaya, Gujarat (IN); Rajesh Aug. 5, 2002 (IN)................................. 699/MUM/2002 Singh Karan, Gujarat (IN); Sunil Aug. 5, 2002 (IN). ... 697/MUM/2002 Sadanand, Gujarat (IN); Vinod Kumar Jan. 22, 2003 (IN)................................... 80/MUM/2003 Gupta, Gujarat (IN) Jan. 22, 2003 (IN)................................... 82/MUM/2003 Correspondence Address: Publication Classification HEDMAN & COSTIGAN P.C. (51) Int. Cl. 1185 AVENUE OF THE AMERICAS A6IK 9/22 (2006.01) NEW YORK, NY 10036 (US) (52) U.S. Cl. .............................................................. 424/468 (22) Filed: May 19, 2005 A dosage form comprising of a high dose, high Solubility active ingredient as modified release and a low dose active ingredient as immediate release where the weight ratio of Related U.S. Application Data immediate release active ingredient and modified release active ingredient is from 1:10 to 1:15000 and the weight of (63) Continuation-in-part of application No. 10/630,446, modified release active ingredient per unit is from 500 mg to filed on Jul. 29, 2003. 1500 mg, a process for preparing the dosage form. Patent Application Publication Feb. 2, 2006 Sheet 1 of 10 US 2006/0024.365A1 FIGURE 1 FIGURE 2 FIGURE 3 Patent Application Publication Feb. 2, 2006 Sheet 2 of 10 US 2006/0024.365A1 FIGURE 4 (a) 7 FIGURE 4 (b) Patent Application Publication Feb. 2, 2006 Sheet 3 of 10 US 2006/0024.365 A1 FIGURE 5 100 ov -- 60 40 20 C 2 4. -

ECO Cup One Step Drug Test Forensic Insert
paranoia, hallucinations, and psychotic behavior. The effects of Amphetamines generally last 2-4 unconsciousness. hours following use, and the drug has a half-life of 4-24 hours in the body. About 30% of Cocaine is often self-administered by nasal inhalation, intravenous injection and free-base Amphetamines are excreted in the urine in unchanged form, with the remainder as hydroxylated smoking. It is excreted in the urine in a short time primarily as Benzoylecgonine. 1.2 and deaminated derivatives. Benzoylecgonine, a major metabolite of cocaine, has a longer biological half-life (5-8 hours) than 2 The ECO CUP™ One Step Drug Test yields a positive result when the concentration of Amphetamine cocaine (0.5-1.5 hours), and can generally be detected for 24-48 hours after cocaine exposure. in urine exceeds 1,000 ng/mL. This is the suggested screening cut-off for positive specimens The ECO CUP™ One Step Drug Test yields a positive result when the concentration of Benzoylecgonine set by the Substance Abuse and Mental Health Services Administration (SAMHSA, USA).3 in urine exceeds 300 ng/mL. This is the suggested screening cut-off for positive specimens set by One Step Drug Test the Substance Abuse and Mental Health Services Administration (SAMHSA, USA). 3 Package Insert for Multi Drug Screen Test Cup AMPHETAMINE (AMP 500) COCAINE (COC 150) This Instruction Sheet is for testing of any combination of the following drugs: See AMPHETAMINE (AMP 1000) for the summary. See COCAINE (COC 300) for the summary. AMP/BAR/BZO/BUP/COC/THC/MTD/mAMP/MDMA/MOR/OPI/OXY/PCP/PPX/TCA/EDDP/6-ACM/ETG The ECO CUP™ One Step Drug Test yields a positive result when the concentration of The ECO CUP™ One Step Drug Test yields a positive result when the concentration of Including Adulterant Tests (Specimen Validity Tests) for: Amphetamine in urine exceeds 500 ng/mL. -

Alltech® Drug Standards for the Forensic, Clinical & Pharmaceutical Industries OH
Alltech® Drug Standards For the Forensic, Clinical & Pharmaceutical Industries OH H3C H H H3C H HO Catalog #505B Our Company Welcome to the Grace's Alltech® Drug Standards Catalog W. R. Grace has manufactured high-quality silica for over 150 years. Grace has been behind the scenes for the past 30 years supplying silica to the chromatography industry. Now we’re in the forefront moving beyond silica, developing and delivering innovative complementary products direct to the customer. Grace Davison Discovery Sciences was founded on Grace’s core strength as a premier manufacturer of differentiated media for SPE, Flash, HPLC, and Process chromatography. This core competency is further enhanced by bringing seven well-known global separations companies together, creating a powerful new single source for all your chromatography needs. A Full Portfolio of Chromatography Products to Support Drug Standards: • HPLC Columns • HPLC Accessories • Flash Products • TLC Products • GC Columns • GC Accessories • SPE and Filtration • Equipment • Syringes • Tubing • Vials For complete details, download the Chromatography Essentials catalog from the Grace web site or contact your customer service representative. Alltech - Part of the Grace Family of Products In 2004, Alltech Associates Inc. was acquired by Grace along with the Alltech® Drug Standards product line. Through investment in research and strategic acquisitions, Grace has expanded our product range and global reach while drawing upon the support of the Grace corporate infrastructure and more than 6000 employees globally to support scientific research and analysis worldwide. With key manufacturing sites in North and South America, Europe, and Asia, plus an extensive international sales and distribution network, separation scientists throughout the world can count on timely delivery and expert local technical service. -

The Sigma1 Protein As a Target for the Non-Genomic Effects of Neuro(Active)Steroids: Molecular, Physiological, and Behavioral Aspects François P
J Pharmacol Sci 100, 93 – 118 (2006) Journal of Pharmacological Sciences ©2006 The Japanese Pharmacological Society Critical Review The Sigma1 Protein as a Target for the Non-genomic Effects of Neuro(active)steroids: Molecular, Physiological, and Behavioral Aspects François P. Monnet1 and Tangui Maurice2,* 1Unité 705 de l’Institut National de la Santé et de la Recherche Médicale, Unité Mixte de Recherche 7157 du Centre National de la Recherche Scientifique, Université de Paris V et VII, Hôpital Lariboisière-Fernand Widal, 2, rue Ambroise Paré, 75475 Paris cedex 10, France 2Unité 710 de l’Institut National de la Santé et de la Recherche Médicale, Ecole Pratique des Hautes Etudes, Université de Montpellier II, cc 105, place Eugène Bataillon, 34095 Montpellier cedex 5, France Received December 15, 2005 Abstract. Steroids synthesized in the periphery or de novo in the brain, so called ‘neuro- steroids’, exert both genomic and nongenomic actions on neurotransmission systems. Through rapid modulatory effects on neurotransmitter receptors, they influence inhibitory and excitatory neurotransmission. In particular, progesterone derivatives like 3α-hydroxy-5α-pregnan-20-one (allopregnanolone) are positive allosteric modulators of the γ-aminobutyric acid type A (GABAA) receptor and therefore act as inhibitory steroids, while pregnenolone sulphate (PREGS) and dehydroepiandrosterone sulphate (DHEAS) are negative modulators of the GABAA receptor and positive modulators of the N-methyl-D-aspartate (NMDA) receptor, therefore acting as excitatory neurosteroids. Some steroids also interact with atypical proteins, the sigma (σ) receptors. Recent studies particularly demonstrated that the σ1 receptor contributes effectively to their pharmaco- logical actions. The present article will review the data demonstrating that the σ1 receptor binds neurosteroids in physiological conditions. -
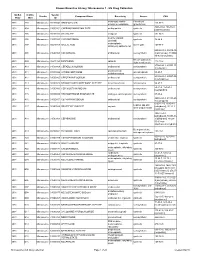
Known Bioactive Library: Microsource 1 - US Drug Collection
Known Bioactive Library: Microsource 1 - US Drug Collection ICCB-L ICCB-L Vendor Vendor Compound Name Bioactivity Source CAS Plate Well ID antifungal, inhibits Penicillium 2091 A03 Microsource 00200046 GRISEOFULVIN 126-07-8 mitosis in metaphase griseofulvum 3505-38-2, 486-16-8 2091 A04 Microsource 01500161 CARBINOXAMINE MALEATE antihistaminic synthetic [carbinoxamine] 2091 A05 Microsource 00200331 SALSALATE analgesic synthetic 552-94-3 muscle relaxant 2091 A06 Microsource 01500162 CARISOPRODOL synthetic 78-44-4 (skeletal) antineoplastic, 2091 A07 Microsource 00210369 GALLIC ACID insect galls 149-91-7 astringent, antibacterial 66592-87-8, 50370-12- 2091 A08 Microsource 01500163 CEFADROXIL antibacterial semisynthetic 2 [anhydrous], 119922- 89-9 [hemihydrate] Rheum palmatum, 2091 A09 Microsource 00211468 DANTHRON cathartic 117-10-2 Xyris semifuscata 27164-46-1, 25953-19- 2091 A10 Microsource 01500164 CEFAZOLIN SODIUM antibacterial semisynthetic 9 [cefazolin] glucocorticoid, 2091 A11 Microsource 00300024 HYDROCORTISONE adrenal glands 50-23-7 antiinflammatory 64485-93-4, 63527-52- 2091 A12 Microsource 01500165 CEFOTAXIME SODIUM antibacterial semisynthetic 6 [cefotaxime] 2091 A13 Microsource 00300029 DESOXYCORTICOSTERONE ACETATE mineralocorticoid adrenocortex 56-47-3 58-71-9, 153-61-7 2091 A14 Microsource 01500166 CEPHALOTHIN SODIUM antibacterial semisynthetic [cephalothin] 2091 A15 Microsource 00300034 TESTOSTERONE PROPIONATE androgen, antineoplastic semisynthetic 57-85-2 24356-60-3, 21593-23- 2091 A16 Microsource 01500167 CEPHAPIRIN SODIUM -

In Vitro Inhibition of Breast Cancer Spheroid-Induced Lymphendothelial
FULL PAPER British Journal of Cancer (2013) 108, 570–578 | doi: 10.1038/bjc.2012.580 Keywords: lymphendothelial intravasation; tumour spheroid; acetohexamide; nifedipin; isoxsuprine; proadifen In vitro inhibition of breast cancer spheroid-induced lymphendothelial defects resembling intravasation into the lymphatic vasculature by acetohexamide, isoxsuprine, nifedipin and proadifen N Kretschy1,8, M Teichmann1,8, S Kopf1, A G Atanasov2, P Saiko3, C Vonach1, K Viola1, B Giessrigl1, N Huttary1, I Raab1, S Krieger1,WJa¨ ger4, T Szekeres3, S M Nijman5, W Mikulits6, V M Dirsch2, H Dolznig7, M Grusch6 and G Krupitza*,1 1Institute of Clinical Pathology, Medical University of Vienna, Waehringer Guertel 18-20, A-1090 Vienna, Austria; 2Department of Pharmacognosy, University of Vienna, Althanstrasse 14, A-1090 Vienna, Austria; 3Clinical Institute of Medical and Chemical Laboratory Diagnostics, Medical University of Vienna, Waehringer Guertel 18-20, A-1090 Vienna, Austria; 4Department for Clinical Pharmacy and Diagnostics, Faculty of Life Sciences, University of Vienna, Althanstrasse 14, A-1090 Vienna, Austria; 5Research Center for Molecular Medicine of the Austrian Academy of Sciences, Lazarettgasse 14, AKH BT 25.3, A-1090 Vienna, Austria; 6Department of Medicine I, Institute of Cancer Research, Medical University of Vienna, Borschkegasse 8a, A-1090 Vienna, Austria and 7Institute of Medical Genetics, Medical University of Vienna, Waehringer Strasse 10, A-1090 Vienna, Austria Background: As metastasis is the prime cause of death from malignancies, there is vibrant interest to discover options for the management of the different mechanistic steps of tumour spreading. Some approved pharmaceuticals exhibit activities against diseases they have not been developed for. In order to discover such activities that might attenuate lymph node metastasis, we investigated 225 drugs, which are approved by the US Food and Drug Administration. -

Forensic Science
Anal. Chem. 2007, 79, 4365-4384 Forensic Science T. A. Brettell* Department of Chemical and Physical Sciences, Cedar Crest College, 100 College Drive, Allentown, Pennsylvania 18104-6196 J. M. Butler Biochemical Science Division, National Institute of Standards and Technology, Gaithersburg, Maryland 20899-8311 J. R. Almirall Department of Chemistry and Biochemistry and International Forensic Research Institute, Florida International University, University Park, Miami, Florida 33199 Review Contents have recently been launched as well: Forensic Science, Medicine, Forensic DNA Analysis 4365 and Pathology (Humana Press) and Forensic Science Interna- Collection, Characterization, Preservation, 4366 tional: Genetics (Elsevier). Our literature survey encompasses the Extraction, and Quantitation of Biological period from January 2005 through December 2006. The format Material selected for this survey divides coverage into three distinct Short Tandem Repeats 4367 areas: forensic DNA analysis, trace evidence, and drugs and Single Nucleotide Polymorphisms 4367 poisons. Within the scope of each of the areas, key articles have Y-Chromosome and X-Chromosome Analysis 4368 Mitochondrial DNA Typing 4369 been selected to describe current forensic science practices in Nonhuman DNA Typing Systems and Microbial 4369 analytical chemistry and to outline relevant forensic science Forensics research interests. In accordance with the policy of the managing DNA Databases, Mass Screens, and Mass 4369 editor, we have strived to keep this review limited to important Disasters articles and to keep our discussions concise and meaningful. Interpretation and Statistical Weight of DNA 4370 Typing Results General Reviews 4370 FORENSIC DNA ANALYSIS Trace Evidence 4370 The literature for forensic DNA analysis has expanded rapidly Petroleum Products and Explosives 4370 in the past few years as various technologies and genetic markers Hairs, Fibers, Glass, and Paint 4371 have been adopted, validated, and examined in numerous popula- Gunshot Residue Analysis 4372 tions around the world. -

Evaluation of a Lc/Ms Method to Screen for Drugs in Post-Mortem Whole Blood Specimens
[ APPLICATION NOTE ] EVALUATION OF A LC/MS METHOD TO SCREEN FOR DRUGS IN POST-MORTEM WHOLE BLOOD SPECIMENS Kevin Shanks1*, Tim Dahn1, Andrea R. Terrell, Ph.D.1, and Jan Bohuslavek, Ph.D.2 1 AIT Laboratories, Indianapolis, IN, 2 Waters Corporation, Milford, MA, USA INTRODUCTION • ChromaLynx™ also produces a list of ‘’candidate’’ components and applies confidence factors to the identification. Toxicological screening of post-mortem whole blood specimens is rou- tinely performed to help determine the cause of death. Traditionally, Retention time data is also used in component identification screening is performed using either GC/MS, immunoassays, or HPLC process which increases confidence in the library search results. The with UV detection. Immunoassays can be cost prohibitive and often results are then displayed in an easy to view browser format. The suffer from cross reactivity . HPLC with UV detection often lacks spec- processed data browser is fully customizable and can contain an ificity and sensitivity. GC/MS requires extensive sample preparation overlayed chromatogram of all functions, spectral information for and is not suitable for thermolabile compounds. An LC/MS approach every component and its corresponding library hit, a list of identi- can potentially overcome many of these limitations and provide a fied candidates, and other relevant information (Figure 1). more thorough screening solution. The aim of the work described in this application note was to compare a new LC/MS screening method to an existing GC/MS method. A key element of the study was to evaluate the efficiency of ChromaLynx™ deconvolution and the library searching software utilized in the LC/MS screening method. -

Y0311141702 WH通用说明书(Forensic)加ETG 2017.03.14
AMPHETAMINE (AMP 1000) 2002 for use in opioid addiction treatment. Buprenorphine was rescheduled from Schedule V to One Step Drug of Abuse Test Amphetamine is a Schedule II controlled substance available by prescription (Dexedrine®) and is Schedule III drug just before FDA approval of Suboxone and Subutex. (Strip, Dipcard, Cassette, Cup) also available on the illicit market. Amphetamines are a class of potent sympathomimetic agents The BUP assay contained within the One Step Drug of Abuse Test yields a positive result when the with therapeutic applications. They are chemically related to the human body's natural concentration of Buprenorphine in urine exceeds 10 ng/mL. catecholamines: epinephrine and norepinephrine. Acute higher doses lead to enhanced Package Insert for Multi Drug Screen Test stimulation of the central nervous system and induce euphoria, alertness, reduced appetite, and a COCAINE (COC 300) sense of increased energy and power. Cardiovascular responses to Amphetamines include Cocaine is a potent central nervous system (CNS) stimulant and a local anesthetic. Initially, it This Instruction Sheet is for testing of any combination of the following drugs: increased blood pressure and cardiac arrhythmias. More acute responses produce anxiety, brings about extreme energy and restlessness while gradually resulting in tremors, over-sensitivity AMP/BAR/BZO/BUP/COC/THC/MTD/mAMP/MDMA/MOR/OPI/OXY/PCP/PPX/TCA/EDDP/6-ACM/COT paranoia, hallucinations, and psychotic behavior. The effects of Amphetamines generally last 2-4 and spasms. In large amounts, cocaine causes fever, unresponsiveness, difficulty in breathing and /K2/KET/FEN/TRA/ETG/ALCO hours following use, and the drug has a half-life of 4-24 hours in the body. -

Synergistic and Antagonistic Drug Interactions in the Treatment
RESEARCH ARTICLE Synergistic and antagonistic drug interactions in the treatment of systemic fungal infections Morgan A Wambaugh, Steven T Denham, Magali Ayala, Brianna Brammer, Miekan A Stonhill, Jessica CS Brown* Division of Microbiology and Immunology, Pathology Department, University of Utah School of Medicine, Salt Lake City, United States Abstract Invasive fungal infections cause 1.6 million deaths annually, primarily in immunocompromised individuals. Mortality rates are as high as 90% due to limited treatments. The azole class antifungal, fluconazole, is widely available and has multi-species activity but only inhibits growth instead of killing fungal cells, necessitating long treatments. To improve treatment, we used our novel high-throughput method, the overlap2 method (O2M) to identify drugs that interact with fluconazole, either increasing or decreasing efficacy. We identified 40 molecules that act synergistically (amplify activity) and 19 molecules that act antagonistically (decrease efficacy) when combined with fluconazole. We found that critical frontline beta-lactam antibiotics antagonize fluconazole activity. A promising fluconazole-synergizing anticholinergic drug, dicyclomine, increases fungal cell permeability and inhibits nutrient intake when combined with fluconazole. In vivo, this combination doubled the time-to-endpoint of mice with Cryptococcus neoformans meningitis. Thus, our ability to rapidly identify synergistic and antagonistic drug interactions can potentially alter the patient outcomes. *For correspondence: Introduction [email protected] Invasive fungal infections are an increasing problem worldwide, contributing to 1.6 million deaths annually (Almeida et al., 2019; Bongomin et al., 2017; Brown et al., 2012). These problematic Competing interests: The infections are difficult to treat for many reasons. Delayed diagnoses, the paucity and toxicity of anti- authors declare that no fungal drugs, and the already immunocompromised state of many patients result in mortality rates competing interests exist. -

I PROPOXYPHENE, NORPROPOXYPHENE, AND
PROPOXYPHENE, NORPROPOXYPHENE, AND PROADIFEN (SKF-525A) ARE MECHANISM-BASED INHIBITORS OF CYP3A4, CYP3A5, AND CYP3A IN HUMAN LIVER MICROSOMES Anna Ruth Riley Submitted to the faculty of the University Graduate School in partial fulfillment of the requirements for the degree Master of Science in the Department of Pharmacology and Toxicology, Indiana University December 2008 i Accepted by the Faculty of Indiana University, in partial fulfillment of the requirements for the degree of Master of Science. ___________________________ Sherry F. Queener, Ph.D., Chair ___________________________ David R. Jones, Ph.D. Master’s Thesis Committee ___________________________ David A. Flockhart, M.D., Ph.D. ___________________________ Lynn R. Willis, Ph.D. ii DEDICATION Special thanks to my family, and friends (especially MK), and MJK who helped me during this journey iii ACKNOWLEDGEMENTS To my committee: Sherry F. Queener David R. Jones David A. Flockhart Lynn R. Willis Eli Lilly and Company, for sponsorship iv ABSTRACT Anna Ruth Riley PROPOXYPHENE, NORPROPOXYPHENE, AND PROADIFEN (SKF-525A) ARE MECHANISM-BASED INHIBITORS OF CYP3A4, CYP3A5, AND CYP3A IN HUMAN LIVER MICROSOMES The purpose of this study is to determine if propoxyphene and norpropoxyphene are mechanism-based (irreversible) inhibitors of CYP3A, and to determine if propoxyphene and norpropoxyphene are reversible inhibitors of CYP3A. Mechanism- based inhibition is a type of irreversible inhibition that results from an inhibitor or its metabolite binding to an enzyme during drug metabolism, which renders the enzyme nonfunctional. Propoxyphene is an analgesic that is frequently prescribed in the United States and Europe. It is metabolized by CYP3A enzymes, and is an irreversible inhibitor of CYP3A4. The major metabolite of propoxyphene is norpropoxyphene, which has not been extensively studied for enzyme inhibition.