1 Linear Least Squares Regression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Ordinary Least Squares 1 Ordinary Least Squares
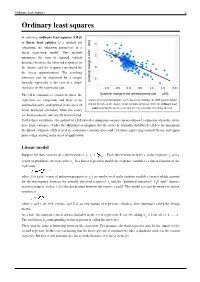
Ordinary least squares 1 Ordinary least squares In statistics, ordinary least squares (OLS) or linear least squares is a method for estimating the unknown parameters in a linear regression model. This method minimizes the sum of squared vertical distances between the observed responses in the dataset and the responses predicted by the linear approximation. The resulting estimator can be expressed by a simple formula, especially in the case of a single regressor on the right-hand side. The OLS estimator is consistent when the regressors are exogenous and there is no Okun's law in macroeconomics states that in an economy the GDP growth should multicollinearity, and optimal in the class of depend linearly on the changes in the unemployment rate. Here the ordinary least squares method is used to construct the regression line describing this law. linear unbiased estimators when the errors are homoscedastic and serially uncorrelated. Under these conditions, the method of OLS provides minimum-variance mean-unbiased estimation when the errors have finite variances. Under the additional assumption that the errors be normally distributed, OLS is the maximum likelihood estimator. OLS is used in economics (econometrics) and electrical engineering (control theory and signal processing), among many areas of application. Linear model Suppose the data consists of n observations { y , x } . Each observation includes a scalar response y and a i i i vector of predictors (or regressors) x . In a linear regression model the response variable is a linear function of the i regressors: where β is a p×1 vector of unknown parameters; ε 's are unobserved scalar random variables (errors) which account i for the discrepancy between the actually observed responses y and the "predicted outcomes" x′ β; and ′ denotes i i matrix transpose, so that x′ β is the dot product between the vectors x and β. -

An Analysis of Random Design Linear Regression
An Analysis of Random Design Linear Regression Daniel Hsu1,2, Sham M. Kakade2, and Tong Zhang1 1Department of Statistics, Rutgers University 2Department of Statistics, Wharton School, University of Pennsylvania Abstract The random design setting for linear regression concerns estimators based on a random sam- ple of covariate/response pairs. This work gives explicit bounds on the prediction error for the ordinary least squares estimator and the ridge regression estimator under mild assumptions on the covariate/response distributions. In particular, this work provides sharp results on the \out-of-sample" prediction error, as opposed to the \in-sample" (fixed design) error. Our anal- ysis also explicitly reveals the effect of noise vs. modeling errors. The approach reveals a close connection to the more traditional fixed design setting, and our methods make use of recent ad- vances in concentration inequalities (for vectors and matrices). We also describe an application of our results to fast least squares computations. 1 Introduction In the random design setting for linear regression, one is given pairs (X1;Y1);:::; (Xn;Yn) of co- variates and responses, sampled from a population, where each Xi are random vectors and Yi 2 R. These pairs are hypothesized to have the linear relationship > Yi = Xi β + i for some linear map β, where the i are noise terms. The goal of estimation in this setting is to find coefficients β^ based on these (Xi;Yi) pairs such that the expected prediction error on a new > 2 draw (X; Y ) from the population, measured as E[(X β^ − Y ) ], is as small as possible. -
![ROBUST LINEAR LEAST SQUARES REGRESSION 3 Bias Term R(F ∗) R(F (Reg)) Has the Order D/N of the Estimation Term (See [3, 6, 10] and References− Within)](https://docslib.b-cdn.net/cover/5087/robust-linear-least-squares-regression-3-bias-term-r-f-r-f-reg-has-the-order-d-n-of-the-estimation-term-see-3-6-10-and-references-within-1085087.webp)
ROBUST LINEAR LEAST SQUARES REGRESSION 3 Bias Term R(F ∗) R(F (Reg)) Has the Order D/N of the Estimation Term (See [3, 6, 10] and References− Within)
The Annals of Statistics 2011, Vol. 39, No. 5, 2766–2794 DOI: 10.1214/11-AOS918 c Institute of Mathematical Statistics, 2011 ROBUST LINEAR LEAST SQUARES REGRESSION By Jean-Yves Audibert and Olivier Catoni Universit´eParis-Est and CNRS/Ecole´ Normale Sup´erieure/INRIA and CNRS/Ecole´ Normale Sup´erieure and INRIA We consider the problem of robustly predicting as well as the best linear combination of d given functions in least squares regres- sion, and variants of this problem including constraints on the pa- rameters of the linear combination. For the ridge estimator and the ordinary least squares estimator, and their variants, we provide new risk bounds of order d/n without logarithmic factor unlike some stan- dard results, where n is the size of the training data. We also provide a new estimator with better deviations in the presence of heavy-tailed noise. It is based on truncating differences of losses in a min–max framework and satisfies a d/n risk bound both in expectation and in deviations. The key common surprising factor of these results is the absence of exponential moment condition on the output distribution while achieving exponential deviations. All risk bounds are obtained through a PAC-Bayesian analysis on truncated differences of losses. Experimental results strongly back up our truncated min–max esti- mator. 1. Introduction. Our statistical task. Let Z1 = (X1,Y1),...,Zn = (Xn,Yn) be n 2 pairs of input–output and assume that each pair has been independently≥ drawn from the same unknown distribution P . Let denote the input space and let the output space be the set of real numbersX R, so that P is a proba- bility distribution on the product space , R. -

Testing for Heteroskedastic Mixture of Ordinary Least 5
TESTING FOR HETEROSKEDASTIC MIXTURE OF ORDINARY LEAST 5. SQUARES ERRORS Chamil W SENARATHNE1 Wei JIANGUO2 Abstract There is no procedure available in the existing literature to test for heteroskedastic mixture of distributions of residuals drawn from ordinary least squares regressions. This is the first paper that designs a simple test procedure for detecting heteroskedastic mixture of ordinary least squares residuals. The assumption that residuals must be drawn from a homoscedastic mixture of distributions is tested in addition to detecting heteroskedasticity. The test procedure has been designed to account for mixture of distributions properties of the regression residuals when the regressor is drawn with reference to an active market. To retain efficiency of the test, an unbiased maximum likelihood estimator for the true (population) variance was drawn from a log-normal normal family. The results show that there are significant disagreements between the heteroskedasticity detection results of the two auxiliary regression models due to the effect of heteroskedastic mixture of residual distributions. Forecasting exercise shows that there is a significant difference between the two auxiliary regression models in market level regressions than non-market level regressions that supports the new model proposed. Monte Carlo simulation results show significant improvements in the model performance for finite samples with less size distortion. The findings of this study encourage future scholars explore possibility of testing heteroskedastic mixture effect of residuals drawn from multiple regressions and test heteroskedastic mixture in other developed and emerging markets under different market conditions (e.g. crisis) to see the generalisatbility of the model. It also encourages developing other types of tests such as F-test that also suits data generating process. -

Bayesian Uncertainty Analysis for a Regression Model Versus Application of GUM Supplement 1 to the Least-Squares Estimate
Home Search Collections Journals About Contact us My IOPscience Bayesian uncertainty analysis for a regression model versus application of GUM Supplement 1 to the least-squares estimate This content has been downloaded from IOPscience. Please scroll down to see the full text. 2011 Metrologia 48 233 (http://iopscience.iop.org/0026-1394/48/5/001) View the table of contents for this issue, or go to the journal homepage for more Download details: IP Address: 129.6.222.157 This content was downloaded on 20/02/2014 at 14:34 Please note that terms and conditions apply. IOP PUBLISHING METROLOGIA Metrologia 48 (2011) 233–240 doi:10.1088/0026-1394/48/5/001 Bayesian uncertainty analysis for a regression model versus application of GUM Supplement 1 to the least-squares estimate Clemens Elster1 and Blaza Toman2 1 Physikalisch-Technische Bundesanstalt, Abbestrasse 2-12, 10587 Berlin, Germany 2 Statistical Engineering Division, Information Technology Laboratory, National Institute of Standards and Technology, US Department of Commerce, Gaithersburg, MD, USA Received 7 March 2011, in final form 5 May 2011 Published 16 June 2011 Online at stacks.iop.org/Met/48/233 Abstract Application of least-squares as, for instance, in curve fitting is an important tool of data analysis in metrology. It is tempting to employ the supplement 1 to the GUM (GUM-S1) to evaluate the uncertainty associated with the resulting parameter estimates, although doing so is beyond the specified scope of GUM-S1. We compare the result of such a procedure with a Bayesian uncertainty analysis of the corresponding regression model. -

Chapter 1 Linear Regression with One Predictor Variable
54 Part I Simple Linear Regression 55 Chapter 1 Linear Regression With One Predictor Variable We look at linear regression models with one predictor variable. 1.1 Relations Between Variables Scatter plots of data are often modelled (approximated, simpli¯ed) by linear or non- linear functions. One \good" way to model the data is by using statistical relations. Whereas functions pass through every point in the scatter plot, statistical relations do not necessarily pass through every point but do follow the systematic pattern of the data. Exercise 1.1 (Functional Relation and Statistical Relation) 1. Linear Function Model: Reading Ability Versus Level of Illumination. Consider the following three linear functions used to model the reading ability versus level of illumination data. y = (14/6)x + 69 y = 1.88x + 74.1 y = 6x + 58 B(7,100) B(7,100) 100 100 B(7,100) 100 y y C(9.90) , C(9.90) , y C(9.90) y y , y ilit ilit b b ilit a 80 a 80 b a 80 g g A(3,76) A(3,76) g A(3,76) in in d d in d ea ea r r ea r 60 60 60 0 2 4 6 8 10 0 2 4 6 8 10 0 2 4 6 8 10 level of illumination, x level of illumination, x level of illumination, x (b) two point BC linear function model (c) least squares linear function model (a) two point AB linear function model Figure 1.1 (Linear Function Models of Reading Ability Versus Level of Illumination) 57 58 Chapter 1. -

Chapter 7 Least Squares Estimation
7-1 Least Squares Estimation Version 1.3 Chapter 7 Least Squares Estimation 7.1. Introduction Least squares is a time-honored estimation procedure, that was developed independently by Gauss (1795), Legendre (1805) and Adrain (1808) and published in the first decade of the nineteenth century. It is perhaps the most widely used technique in geophysical data analysis. Unlike maximum likelihood, which can be applied to any problem for which we know the general form of the joint pdf, in least squares the parameters to be estimated must arise in expressions for the means of the observations. When the parameters appear linearly in these expressions then the least squares estimation problem can be solved in closed form, and it is relatively straightforward to derive the statistical properties for the resulting parameter estimates. One very simple example which we will treat in some detail in order to illustrate the more general problem is that of fitting a straight line to a collection of pairs of observations (xi, yi) where i = 1, 2, . , n. We suppose that a reasonable model is of the form y = β0 + β1x, (1) and we need a mechanism for determining β0 and β1. This is of course just a special case of many more general problems including fitting a polynomial of order p, for which one would need to find p + 1 coefficients. The most commonly used method for finding a model is that of least squares estimation. It is supposed that x is an independent (or predictor) variable which is known exactly, while y is a dependent (or response) variable. -

Week 6: Linear Regression with Two Regressors
Week 6: Linear Regression with Two Regressors Brandon Stewart1 Princeton October 17, 19, 2016 1These slides are heavily influenced by Matt Blackwell, Adam Glynn and Jens Hainmueller. Stewart (Princeton) Week 6: Two Regressors October 17, 19, 2016 1 / 132 Where We've Been and Where We're Going... Last Week I mechanics of OLS with one variable I properties of OLS This Week I Monday: F adding a second variable F new mechanics I Wednesday: F omitted variable bias F multicollinearity F interactions Next Week I multiple regression Long Run I probability inference regression ! ! Questions? Stewart (Princeton) Week 6: Two Regressors October 17, 19, 2016 2 / 132 1 Two Examples 2 Adding a Binary Variable 3 Adding a Continuous Covariate 4 Once More With Feeling 5 OLS Mechanics and Partialing Out 6 Fun With Red and Blue 7 Omitted Variables 8 Multicollinearity 9 Dummy Variables 10 Interaction Terms 11 Polynomials 12 Conclusion 13 Fun With Interactions Stewart (Princeton) Week 6: Two Regressors October 17, 19, 2016 3 / 132 Why Do We Want More Than One Predictor? Summarize more information for descriptive inference Improve the fit and predictive power of our model Control for confounding factors for causal inference 2 Model non-linearities (e.g. Y = β0 + β1X + β2X ) Model interactive effects (e.g. Y = β0 + β1X + β2X2 + β3X1X2) Stewart (Princeton) Week 6: Two Regressors October 17, 19, 2016 4 / 132 Example 1: Cigarette Smokers and Pipe Smokers Stewart (Princeton) Week 6: Two Regressors October 17, 19, 2016 5 / 132 Example 1: Cigarette Smokers and Pipe Smokers Consider the following example from Cochran (1968). -

Fast Model-Fitting of Bayesian Variable Selection Regression Using The
Fast model-fitting of Bayesian variable selection regression using the iterative complex factorization algorithm Quan Zhou ∗,z and Yongtao Guan y,x Abstract. Bayesian variable selection regression (BVSR) is able to jointly ana- lyze genome-wide genetic datasets, but the slow computation via Markov chain Monte Carlo (MCMC) hampered its wide-spread usage. Here we present a novel iterative method to solve a special class of linear systems, which can increase the speed of the BVSR model-fitting tenfold. The iterative method hinges on the complex factorization of the sum of two matrices and the solution path resides in the complex domain (instead of the real domain). Compared to the Gauss-Seidel method, the complex factorization converges almost instantaneously and its error is several magnitude smaller than that of the Gauss-Seidel method. More impor- tantly, the error is always within the pre-specified precision while the Gauss-Seidel method is not. For large problems with thousands of covariates, the complex fac- torization is 10 { 100 times faster than either the Gauss-Seidel method or the direct method via the Cholesky decomposition. In BVSR, one needs to repetitively solve large penalized regression systems whose design matrices only change slightly be- tween adjacent MCMC steps. This slight change in design matrix enables the adaptation of the iterative complex factorization method. The computational in- novation will facilitate the wide-spread use of BVSR in reanalyzing genome-wide association datasets. Keywords: Cholesky decomposition, exchange algorithm, fastBVSR, Gauss-Seidel method, heritability. 1 Introduction Bayesian variable selection regression (BVSR) can jointly analyze genome-wide genetic data to produce the posterior probability of association for each covariate and estimate hyperparameters such as heritability and the number of covariates that have nonzero effects (Guan and Stephens, 2011). -

Chapter 14. Linear Least Squares
Serik Sagitov, Chalmers and GU, March 9, 2016 Chapter 14. Linear least squares 1 Simple linear regression model A linear model for the random response Y = Y (x) to an independent variable X = x. For a given set of values (x1; : : : ; xn) of the independent variable put Yi = β0 + β1xi + i; i = 1; : : : ; n; 2 assuming that the noise vector (1; : : : ; n) has independent N(0,σ ) random components. Given the 2 data (y1; : : : ; yn), the model is characterised by the likelihood function of three parameters β0, β1, σ n 2 S(β ,β ) 2 Y 1 n (yi − β0 − β1xi) o −n=2 −n − 0 1 L(β ; β ; σ ) = p exp − = (2π) σ e 2σ2 ; 0 1 2σ2 i=1 2πσ Pn 2 where S(β0; β1) = i=1(yi − β0 − β1xi) . Observe that n −1 −1 X 2 2 2 2 2 n S(β0; β1) = n (yi − β0 − β1xi) = β0 + 2β0β1x¯ − 2β0y¯ − 2β1xy + β1 x + y : i=1 Least squares estimates Regression lines: true y = β0 + β1x and fitted y = b0 + b1x. We want to find (b0; b1) such that the observed responses yi are approximated by the predicted responsesy ^i = b0 + b1xi in an optimal way. P 2 Least squares method: find (b0; b1) minimising the sum of squares S(b0; b1) = (yi − y^i) . From @S=@b0 = 0 and @S=@b1 = 0 we get the so-called Normal Equations: 8 xy − x¯y¯ rsy b0 + b1x¯ =y ¯ < b1 = = 2 2 2 implying x − x¯ sx b0x¯ + b1x = xy : b0 =y ¯ − b1x¯ The least square regression line y = b + b x takes the form y =y ¯ + r sy (x − x¯): 0 1 sx 2 1 P 2 2 1 P 2 sample variances sx = n−1 (xi − x¯) , sy = n−1 (yi − y¯) , 1 P sample covariance sxy = n−1 (xi − x¯)(yi − y¯), sample correlation coefficient r = sxy . -

Skedastic: Heteroskedasticity Diagnostics for Linear Regression
Package ‘skedastic’ June 14, 2021 Type Package Title Heteroskedasticity Diagnostics for Linear Regression Models Version 1.0.3 Description Implements numerous methods for detecting heteroskedasticity (sometimes called heteroscedasticity) in the classical linear regression model. These include a test based on Anscombe (1961) <https://projecteuclid.org/euclid.bsmsp/1200512155>, Ramsey's (1969) BAMSET Test <doi:10.1111/j.2517-6161.1969.tb00796.x>, the tests of Bickel (1978) <doi:10.1214/aos/1176344124>, Breusch and Pagan (1979) <doi:10.2307/1911963> with and without the modification proposed by Koenker (1981) <doi:10.1016/0304-4076(81)90062-2>, Carapeto and Holt (2003) <doi:10.1080/0266476022000018475>, Cook and Weisberg (1983) <doi:10.1093/biomet/70.1.1> (including their graphical methods), Diblasi and Bowman (1997) <doi:10.1016/S0167-7152(96)00115-0>, Dufour, Khalaf, Bernard, and Genest (2004) <doi:10.1016/j.jeconom.2003.10.024>, Evans and King (1985) <doi:10.1016/0304-4076(85)90085-5> and Evans and King (1988) <doi:10.1016/0304-4076(88)90006-1>, Glejser (1969) <doi:10.1080/01621459.1969.10500976> as formulated by Mittelhammer, Judge and Miller (2000, ISBN: 0-521-62394-4), Godfrey and Orme (1999) <doi:10.1080/07474939908800438>, Goldfeld and Quandt (1965) <doi:10.1080/01621459.1965.10480811>, Harrison and McCabe (1979) <doi:10.1080/01621459.1979.10482544>, Harvey (1976) <doi:10.2307/1913974>, Honda (1989) <doi:10.1111/j.2517-6161.1989.tb01749.x>, Horn (1981) <doi:10.1080/03610928108828074>, Li and Yao (2019) <doi:10.1016/j.ecosta.2018.01.001> -

Instrumental Variable Analysis in Epidemiologic Studies: An
tica Anal eu yt c ic a a m A r c a Uddin et al., Pharm Anal Acta 2015, 6:4 t h a P Pharmaceutica DOI: 10.4172/2153-2435.1000353 Analytica Acta ISSN: 2153-2435 Review Article Open Access Instrumental Variable Analysis in Epidemiologic Studies: An Overview of the Estimation Methods Uddin MJ1, Groenwold RH1,2, Ton de Boer1, Belitser SV1, Roes KC2 and Klungel OH1,2* 1Devision of Pharmacoepidemiology and Clinical Pharmacology, Utrecht Institute for Pharmaceutical Sciences, University of Utrecht, Utrecht, The Netherlands 2Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht, The Netherlands Abstract Instrumental variables (IV)analysis seems an attractive method to control for unmeasured confounding in observational epidemiological studies. Here, we provide an overview of the estimation methods of IVanalysis and indicate their possible advantages and limitations.We found that two-stage least squares is the method of first choice if exposure and outcome are both continuous and show a linear relation. In case of a nonlinear relation, two-stage residual inclusion may be a suitable alternative. In settings with binary outcomes as well as nonlinear relations between exposure and outcome, generalized method of moments (GMM), structural mean models (SMM), and bivariate probit models perform well, yet GMM and SMM are generally more robust. The standard errors of the IVestimate can be estimated using a robust or bootstrap method. All estimation methods are prone to bias when the IVassumptions are violated. Researchers should be aware of the underlying assumptions of the estimation methods as well as the key assumptions of the IVwhen interpreting the exposure effects estimated through IV analysis.