Metabolism of Hydrogen Sulfide (H2S) and Production of Reactive Sulfur Species (RSS) by Superoxide Dismutase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Five Tyrosines and Two Serines in Human Albumin Are Labeled by the Organophosphorus Agent FP-Biotin
View metadata, citation and similar papers at core.ac.uk brought to you by CORE Chem. Res. Toxicol. 2008, 21, 1787–1794 1787 provided by PubMed Central Five Tyrosines and Two Serines in Human Albumin Are Labeled by the Organophosphorus Agent FP-Biotin Shi-Jian Ding,† John Carr,† James E. Carlson,‡ Larry Tong,§ Weihua Xue,§ Yifeng Li,† Lawrence M. Schopfer,§ Bin Li,§ Florian Nachon,| Oluwatoyin Asojo,† Charles M. Thompson,⊥ Steven H. Hinrichs,† Patrick Masson,| and Oksana Lockridge*,§ Department of Pathology and Microbiology, UniVersity of Nebraska Medical Center, Omaha, Nebraska 68198, Applied Biosystems, Framingham, Massachusetts 01701, Eppley Institute, UniVersity of Nebraska Medical Center, Omaha, Nebraska 68198, Centre de Recherches d SerVice de Sante´ des Arme´es, Unite´ d’Enzymologie, BP87, 38702 La Tronche Cedex, France, and Department of Biomedical and Pharmaceutical Sciences, UniVersity of Montana, Missoula, Montana 59812 ReceiVed April 23, 2008 Tyrosine 411 of human albumin is an established site for covalent attachment of 10-fluoroethoxyphosphinyl- N-biotinamidopentyldecanamide (FP-biotin), diisopropylfluorophosphate, chlorpyrifos oxon, soman, sarin, and dichlorvos. This work investigated the hypothesis that other residues in albumin could be modified by organophosphorus agents (OP). Human plasma was aggressively treated with FP-biotin; plasma proteins were separated into high and low abundant portions using a proteome partitioning antibody kit, and the proteins were digested with trypsin. The FP-biotinylated tryptic peptides were isolated by binding to monomeric avidin beads. The major sites of covalent attachment identified by mass spectrometry were Y138, Y148, Y401, Y411, Y452, S232, and S287 of human albumin. Prolonged treatment of pure human albumin with chlorpyrifos oxon labeled Y138, Y150, Y161, Y401, Y411, and Y452. -

Anaerobic Degradation of Methanethiol in a Process for Liquefied Petroleum Gas (LPG) Biodesulfurization
Anaerobic degradation of methanethiol in a process for Liquefied Petroleum Gas (LPG) biodesulfurization Promotoren Prof. dr. ir. A.J.H. Janssen Hoogleraar in de Biologische Gas- en waterreiniging Prof. dr. ir. A.J.M. Stams Persoonlijk hoogleraar bij het laboratorium voor Microbiologie Copromotor Prof. dr. ir. P.N.L. Lens Hoogleraar in de Milieubiotechnologie UNESCO-IHE, Delft Samenstelling promotiecommissie Prof. dr. ir. R.H. Wijffels Wageningen Universiteit, Nederland Dr. ir. G. Muyzer TU Delft, Nederland Dr. H.J.M. op den Camp Radboud Universiteit, Nijmegen, Nederland Prof. dr. ir. H. van Langenhove Universiteit Gent, België Dit onderzoek is uitgevoerd binnen de onderzoeksschool SENSE (Socio-Economic and Natural Sciences of the Environment) Anaerobic degradation of methanethiol in a process for Liquefied Petroleum Gas (LPG) biodesulfurization R.C. van Leerdam Proefschrift ter verkrijging van de graad van doctor op gezag van de rector magnificus van Wageningen Universiteit Prof. dr. M.J. Kropff in het openbaar te verdedigen op maandag 19 november 2007 des namiddags te vier uur in de Aula Van Leerdam, R.C., 2007. Anaerobic degradation of methanethiol in a process for Liquefied Petroleum Gas (LPG) biodesulfurization. PhD-thesis Wageningen University, Wageningen, The Netherlands – with references – with summaries in English and Dutch ISBN: 978-90-8504-787-2 Abstract Due to increasingly stringent environmental legislation car fuels have to be desulfurized to levels below 10 ppm in order to minimize negative effects on the environment as sulfur-containing emissions contribute to acid deposition (‘acid rain’) and to reduce the amount of particulates formed during the burning of the fuel. Moreover, low sulfur specifications are also needed to lengthen the lifetime of car exhaust catalysts. -

Aegls Brochure
4.85 5 5 About the Board on Environmental Studies and Toxicology The Board on Environmental Studies and Toxicology addresses Types of Chemicals Covered in the AEGLs Series environmental pollution problems affecting human health, human impacts on the environment, and the assessment and management of risks to AEGLs values for the chemicals listed below were published in the first human health and the environment. The board’s reports answer questions six volumes of the AEGLs series. AEGLs for additional chemicals will about air and water pollution; solid and hazardous waste; toxicology; continue to be published in subsequent volumes. epidemiology; risk assessment; applied ecology; natural resources; and environmental engineering, economics, law, and policy. Allylamine Hydrogen fluoride Ammonia Iron pentacarbonyl Aniline Methyl hydrazine Arsine Methyl isocyanate About NRC Reports from the National Academies Protecting Chlorine Nerve agents GA [tabun], The National Academies, through its National Research Council reports, Chlorine dioxide GB [sarin], GD [soman], GF, provides a unique public service by working outside the framework of Chlorine trifluoride and VX the Public and government to ensure independent, expert advice on matters of science, Crotonaldehyde Nickel carbonyl technology, and medicine. Today, the National Academies include three Cyclohexylamine Phosgene honorary societies that elect new members to their ranks each year- Diborane Phosphine Emergency the National Academy of Sciences, the National Academy of Engineering, 1,1-Dichloro-1-fluoroethane Propylene glycol dinitrate and the Institute of Medicine-and the National Research Council, the (HCFC-141B) Sulfur mustard operating arm that conducts the bulk of the institution’s Dimethylhydrazine 1,1,1,2-Tetrafluoroethane Workers science-policy and technical work. -

Chemical Name Federal P Code CAS Registry Number Acutely
Acutely / Extremely Hazardous Waste List Federal P CAS Registry Acutely / Extremely Chemical Name Code Number Hazardous 4,7-Methano-1H-indene, 1,4,5,6,7,8,8-heptachloro-3a,4,7,7a-tetrahydro- P059 76-44-8 Acutely Hazardous 6,9-Methano-2,4,3-benzodioxathiepin, 6,7,8,9,10,10- hexachloro-1,5,5a,6,9,9a-hexahydro-, 3-oxide P050 115-29-7 Acutely Hazardous Methanimidamide, N,N-dimethyl-N'-[2-methyl-4-[[(methylamino)carbonyl]oxy]phenyl]- P197 17702-57-7 Acutely Hazardous 1-(o-Chlorophenyl)thiourea P026 5344-82-1 Acutely Hazardous 1-(o-Chlorophenyl)thiourea 5344-82-1 Extremely Hazardous 1,1,1-Trichloro-2, -bis(p-methoxyphenyl)ethane Extremely Hazardous 1,1a,2,2,3,3a,4,5,5,5a,5b,6-Dodecachlorooctahydro-1,3,4-metheno-1H-cyclobuta (cd) pentalene, Dechlorane Extremely Hazardous 1,1a,3,3a,4,5,5,5a,5b,6-Decachloro--octahydro-1,2,4-metheno-2H-cyclobuta (cd) pentalen-2- one, chlorecone Extremely Hazardous 1,1-Dimethylhydrazine 57-14-7 Extremely Hazardous 1,2,3,4,10,10-Hexachloro-6,7-epoxy-1,4,4,4a,5,6,7,8,8a-octahydro-1,4-endo-endo-5,8- dimethanonaph-thalene Extremely Hazardous 1,2,3-Propanetriol, trinitrate P081 55-63-0 Acutely Hazardous 1,2,3-Propanetriol, trinitrate 55-63-0 Extremely Hazardous 1,2,4,5,6,7,8,8-Octachloro-4,7-methano-3a,4,7,7a-tetra- hydro- indane Extremely Hazardous 1,2-Benzenediol, 4-[1-hydroxy-2-(methylamino)ethyl]- 51-43-4 Extremely Hazardous 1,2-Benzenediol, 4-[1-hydroxy-2-(methylamino)ethyl]-, P042 51-43-4 Acutely Hazardous 1,2-Dibromo-3-chloropropane 96-12-8 Extremely Hazardous 1,2-Propylenimine P067 75-55-8 Acutely Hazardous 1,2-Propylenimine 75-55-8 Extremely Hazardous 1,3,4,5,6,7,8,8-Octachloro-1,3,3a,4,7,7a-hexahydro-4,7-methanoisobenzofuran Extremely Hazardous 1,3-Dithiolane-2-carboxaldehyde, 2,4-dimethyl-, O- [(methylamino)-carbonyl]oxime 26419-73-8 Extremely Hazardous 1,3-Dithiolane-2-carboxaldehyde, 2,4-dimethyl-, O- [(methylamino)-carbonyl]oxime. -

Inorganic Syntheses
INORGANIC SYNTHESES Volume 27 .................... ................ Board of Directors JOHN P. FACKLER, JR. Texas A&M University BODlE E. DOUGLAS University of Pittsburgh SMITH L. HOLT, JR. Oklahoma State Uniuersity JAY H. WORRELL University of South Florida RUSSELL N. GRIMES University of Virginia ROBERT J. ANGELIC1 Iowa State University Future Volumes 28 ROBERT J. ANGELIC1 Iowa State University 29 RUSSELL N. GRIMES University of Virginia 30 LEONARD V. INTERRANTE Rensselaer Polytechnic Institute 31 ALLEN H. COWLEY University of Texas, Austin 32 MARCETTA Y. DARENSBOURG Texas A&M University International Associates MARTIN A. BENNETT Australian National University, Canberra FAUSTO CALDERAZZO University of Pisa E. 0. FISCHER Technical University. Munich JACK LEWIS Cambridge University LAMBERTO MALATESTA University of Milan RENE POILBLANC University of Toulouse HERBERT W. ROESKY University of Gottingen F. G. A. STONE University of Bristol GEOFFREY WILKINSON Imperial College of Science and Technology. London AKlO YAMAMOTO Tokyo Institute 01 Technology. Yokohama Editor-in-Chief ALVIN P. GINSBERG INORGANIC SYNTHESES Volume 27 A Wiley-Interscience Publication JOHN WILEY & SONS New York Chichester Brisbane Toronto Singapore A NOTE TO THE READER This book has been electronically reproduced from digital idormation stored at John Wiley h Sons, Inc. We are phased that the use of this new technology will enable us to keep works of enduring scholarly value in print as long as there is a reasonable demand for them. The content of this book is identical to previous printings. Published by John Wiley & Sons, Inc. Copyright $? 1990 Inorganic Syntheses, Inc. All rights reserved. Published simultaneously in Canada. Reproduction or translation of any part of this work beyond that permitted by Section 107 or 108 of the 1976 United States Copyright Act without the permission of the copyright owner is unlawful. -

Polymer Chemistry
Polymer Chemistry View Article Online PAPER View Journal | View Issue Solution processible hyperbranched inverse- vulcanized polymers as new cathode materials Cite this: Polym. Chem., 2015, 6, 973 in Li–S batteries† Yangyang Wei,a Xiang Li,b Zhen Xu,a Haiyan Sun,a Yaochen Zheng,a,c Li Peng,a Zheng Liu,a Chao Gao*a and Mingxia Gao*b Soluble inverse-vulcanized hyperbranched polymers (SIVHPs) were synthesized via thiol–ene addition of polymeric sulfur (S8) radicals to 1,3-diisopropenylbenzene (DIB). Benefiting from their branched molecular architecture, SIVHPs presented excellent solubility in polar organic solvents with an ultrahigh concen- tration of 400 mg mL−1. After end-capping by sequential click chemistry of thiol–ene and Menschutkin quaternization reactions, we obtained water soluble SIVHPs for the first time. The sulfur-rich SIVHPs were employed as solution processible cathode-active materials for Li–S batteries, by facile fluid infiltration into conductive frameworks of graphene-based ultralight aerogels (GUAs). The SIVHPs-based cells showed high initial specific capacities of 1247.6 mA h g−1 with 400 charge–discharge cycles. The cells also demonstrated an excellent rate capability and a considerable depression of shuttle effect with stable cou- Received 24th September 2014, lombic efficiency of around 100%. The electrochemical performance of SIVHP in Li–S batteries over- Accepted 14th October 2014 whelmed the case of neat sulfur, due to the chemical fixation of sulfur. The combination of high DOI: 10.1039/c4py01055h solubility, structure flexibility, and superior electrochemical performance opens a door for the promising www.rsc.org/polymers application of SIVHPs. -

Table II. EPCRA Section 313 Chemical List for Reporting Year 2017 (Including Toxic Chemical Categories)
Table II. EPCRA Section 313 Chemical List For Reporting Year 2017 (including Toxic Chemical Categories) Individually listed EPCRA Section 313 chemicals with CAS numbers are arranged alphabetically starting on page II-3. Following the alphabetical list, the EPCRA Section 313 chemicals are arranged in CAS number order. Covered chemical categories follow. Note: Chemicals may be added to or deleted from the list. The Emergency Planning and Community Right-to-Know Call Center or the TRI-Listed Chemicals website will provide up-to-date information on the status of these changes. See section B.3.c of the instructions for more information on the de minimis % limits listed below. There are no de minimis levels for PBT chemicals since the de minimis exemption is not available for these chemicals (an asterisk appears where a de minimis limit would otherwise appear in Table II). However, for purposes of the supplier notification requirement only, such limits are provided in Appendix C. Chemical Qualifiers Certain EPCRA Section 313 chemicals listed in Table II have parenthetic “qualifiers.” These qualifiers indicate that these EPCRA Section 313 chemicals are subject to the section 313 reporting requirements if manufactured, processed, or otherwise used in a specific form or when a certain activity is performed. An EPCRA Section 313 chemical that is listed without a qualifier is subject to reporting in all forms in which it is manufactured, processed, and otherwise used. The following chemicals are reportable only if they are manufactured, processed, or otherwise used in the specific form(s) listed below: Chemical/ Chemical Category CAS Number Qualifier Aluminum (fume or dust) 7429-90-5 Only if it is a fume or dust form. -
Ttl Iii26 (C) 28
USOO6610977B2 (12) United States Patent (10) Patent No.: US 6,610,977 B2 Megerle (45) Date of Patent: Aug. 26, 2003 (54) SECURITY SYSTEM FOR NBC-SAFE 5,692,029 A 11/1997 Husseiny et al. BUILDING 5,692,446 A 12/1997 Becker et al. 5,866.430 A 2/1999 Grow (75) Inventor: Clifford Megerle, Manassas, VA (US) 5,915,268 A 6/1999 Linker et al. 5,965,882 A 10/1999 Megerle et al. (73) Assignee: Lockheed Martin Corporation, 6,073,499 A 6/2000 Settles Bethesda, MD (US) 6,100,698 A 8/2000 Megerle et al. OTHER PUBLICATIONS * ) Notice: Subject to anyy disclaimer, the term of this patent is extended or adjusted under 35 U.S. Pub: 2003/0009661 A1, “Security System and Method U.S.C. 154(b) by 53 days. of Security Service Business”, Tsutsumi et al. (Jan. 9, 2003 ).* (21) Appl. No.: 09/969,050 * cited by examiner (22) Filed: Oct. 1, 2001 Primary Examiner John R. Lee O O ASSistant Examiner Zia R. Hashmi (65)65 PriorO PublicationCaO Data (74) Attorney, Agent, or Firm-Simpson & Simpson, PLLC US 2003/0085348 A1 May 8, 2003 (57) ABSTRACT (51) Int. Cl." ................................................ G01N 23/00 (52) U.S. Cl. ........................ 250,287: 250/286, 73,232. A method and apparatus for Screening an object for the 73/28.01, 73/23; 7323,36. 37s'ss. 37ss6. presence of an explosive, chemical warfare agent, biological s s s 37s?is6 warfare agent, drug, metal, weapon, and/or radioactive (58) Field of Search 250/287, 286; material. -

SUMMARY of PARTICULARLY HAZARDOUS SUBSTANCES (By
SUMMARY OF PARTICULARLY HAZARDOUS SUBSTANCES (by alpha) Key: SC -- Select Carcinogens RT -- Reproductive Toxins AT -- Acute Toxins SA -- Readily Absorbed Through the Skin DHS -- Chemicals of Interest Revised: 11/2012 ________________________________________________________ ___________ _ _ _ _ _ _ _ _ _ _ _ ||| | | | CHEMICAL NAME CAS # |SC|RT| AT | SA |DHS| ________________________________________________________ ___________ | _ | _ | _ | _ | __ | | | | | | | 2,4,5-T 000093-76-5 | | x | | x | | ABRIN 001393-62-0 | | | x | | | ACETALDEHYDE 000075-07-0 | x | | | | | ACETAMIDE 000060-35-5 | x | | | | | ACETOHYDROXAMIC ACID 000546-88-3 ||x| | x | | ACETONE CYANOHYDRIN, STABILIZED 000075-86-5 | | | x | | x | ACETYLAMINOFLUORENE,2- 000053-96-3 | x | | | | | ACID MIST, STRONG INORGANIC 000000-00-0 | x | | | | | ACROLEIN 000107-02-8 | | x | x | x | | ACRYLAMIDE 000079-06-1 | x | x | | x | | ACRYLONITRILE 000107-13-1 | x | x | x | x | | ACTINOMYCIN D 000050-76-0 ||x| | x | | ADIPONITRILE 000111-69-3 | | | x | | | ADRIAMYCIN 023214-92-8 | x | | | | | AFLATOXIN B1 001162-65-8 | x | | | | | AFLATOXIN M1 006795-23-9 | x | | | | | AFLATOXINS 001402-68-2 | x | | x | | | ALL-TRANS RETINOIC ACID 000302-79-4 | | x | | x | | ALPRAZOMAN 028981-97-7 | | x | | x | | ALUMINUM PHOSPHIDE 020859-73-8 | | | x | | x | AMANTADINE HYDROCHLORIDE 000665-66-7 | | x | | x | | AMINO-2,4-DIBROMOANTHRAQUINONE 000081-49-2 | x | | | | | AMINO-2-METHYLANTHRAQUINONE, 1- 000082-28-0 | x | | | | | AMINO-3,4-DIMETHYL-3h-IMIDAZO(4,5f)QUINOLINE,2- 077094-11-2 | x | | | | | AMINO-3,8-DIMETHYL-3H-IMIDAZO(4,5-f)QUINOXALINE, -

National Advisory Committee for Acute Exposure Guideline Levels for Hazardous Substances Report Run Date: 09/26/2021 04:22:31 PM 1
2008 Current Fiscal Year Report: National Advisory Committee for Acute Exposure Guideline Levels for Hazardous Substances Report Run Date: 09/26/2021 04:22:31 PM 1. Department or Agency 2. Fiscal Year Environmental Protection Agency 2008 3b. GSA 3. Committee or Subcommittee Committee No. National Advisory Committee for Acute Exposure Guideline Levels 2073 for Hazardous Substances 4. Is this New During Fiscal 5. Current 6. Expected Renewal 7. Expected Term Year? Charter Date Date No 11/02/2007 11/02/2009 8a. Was Terminated During 8b. Specific Termination 8c. Actual Term FiscalYear? Authority Date No 9. Agency Recommendation for Next10a. Legislation Req to 10b. Legislation FiscalYear Terminate? Pending? Continue 11. Establishment Authority Agency Authority 12. Specific Establishment 13. Effective 14. Commitee 14c. Authority Date Type Presidential? AGEN 09/28/1995 Continuing No 15. Description of Committee Scientific Technical Program Advisory Board 16a. Total Number of Reports 16 16b. Report Date Report Title 10/01/2007 Final Acute Exposure Guideline Levels for Chlorine Dioxide 10/01/2007 Final Acute Exposure Guideline Levels for Chlorine Trifluoride 10/01/2007 Final Acute Exposure Guideline Levels for Cyclohexylamine 10/01/2007 Final Acute Exposure Guideline Levels for Ethylenediamine 10/01/2007 Final Acute Exposure Guideline Levels for Hydrofluoroether-7100 10/01/2007 Final Acute Exposure Guideline Levels for Tetranitromethane 04/01/2008 Final Acute Exposure Guideline Levels for Allylamine 04/01/2008 Final Acute Exposure Guideline Levels -

Department of Homeland Security (Dhs) Appendix a Chemicals of Interest (Coi)
DEPARTMENT OF HOMELAND SECURITY (DHS) APPENDIX A CHEMICALS OF INTEREST (COI) Acetaldehyde Bromine trifluoride Acetone Cyanohydrin, stabilized Bromothrifluorethylene Acetyl bromide 1,3-Butadiene Acetyl chloride Butane Acetyl iodide Butene Acetylene 1-Butene Acrolein 2-Butene Acrylonitrile 2-Butene-cis Acrylyl chloride 2-Butene-trans Allyl alcohol Butyltrichlorosilane Allylamine Calcium hydrosulfite Allyltrichlorosilane, stabilized Calcium phosphide Aluminum (powder) Carbon disulfide Aluminum Bromide, anhydrous Carbon oxysulfide Aluminum Chloride, anhydrous Carbonyl fluoride Aluminum phosphide Carbonyl sulfide Ammonia (anhydrous) Chlorine Ammonia (conc 20% or greater) Chlorine dioxide Ammonium nitrate, note #1 Chlorine monoxide Ammonium nitrate, note #2 Chlorine pentafluoride Ammonium perchlorate Chlorine trifluoride Ammonium picrate Chloroacetyl chloride Amyltrichlorosilane 2-Chloroethylchloro-methylsulfide Antimony pentafluoride Chloroform Arsenic trichloride Chloromethyl ether Arsine Chloromethyl methyl ether Barium azide 1-Chloropropylene 1,4-Bis(2-chloroethylthio)-n-butane 2-Chloropropylene Bis(2-chloroethylthio) methane Chlorosarin Bis(2-chloroethylthiomethyl)ether Chlorosoman 1,5-Bis(2-chloroethylthio)-n-pentane Chlorosulfonic acid 1,3-Bis(2-chloroethylthio)-n-propane Chromium oxychloride Boron tribromide Crotonaldehyde Boron trichloride Crotonaldehyde, (E)- Boron trifluoride Cyanogen Boron trifluoride compound with methyl Cyanogen chloride ether (1:1) Cyclohexylamine Bromine Cyclohexyltrichlorosilane Bromine chloride Cyclopropane -
Homeland Security List
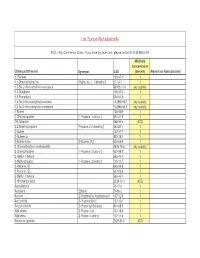
List Sorted Alphabetically ACG = Any Commercial Grade. If you have any questions, please contact EHS @ 898-5126. Minimum Concentration Chemical Of Interest Synonym CAS (percent) Amount on Hand (pounds) 1- Pentene 109-67-1 1 1,1-Dimethylhydrazine [Hydrazine, 1, 1-dimethyl-] 57-14-7 1 1,3-Bis(2-chloroethylthio)-n-propane 63905-10-2 any quantity 1,3-Butadiene 106-99-0 1 1,3-Pentadiene 504-60-9 1 1,4-Bis(2-chloroethylthio)-n-butane 142868-93-7 any quantity 1,5-Bis(2-chloroethylthio)-n-pentane 142868-94-8 any quantity 1-Butene 106-98-9 1 1-Chloropropylene [1-Propene, 1-chloro-] 590-21-6 1 1H-Tetrazole 288-94-8 ACG 2,2-Dimethylpropane [Propane, 2,2-dimethyl-] 463-82-1 1 2-Butene 107-01-7 1 2-Butene-cis 590-18-1 1 2-Butene-trans [2-Butene, (E)] 624-64-6 1 2-Chloroethylchloro-methylsulfide 2625-76-5 any quantity 2-Chloropropylene [1-Propene, 2-chloro-] 557-98-2 1 2-Methyl-1-butene 563-46-2 1 2-Methylpropene [1-Propene, 2-methyl-] 115-11-7 1 2-Pentene, (E)- 646-04-8 1 2-Pentene, (Z)- 627-20-3 1 3-Methyl-1-butene 563-45-1 1 5-Nitrobenzotriazol 2338-12-7 ACG Acetaldehyde 75-07-0 1 Acetylene [Ethyne] 74-86-2 1 Acrolein [2-Propenal] or Acrylaldehyde 107-02-8 1 Acrylonitrile [2-Propenenitrile] 107-13-1 1 Acrylyl chloride [2-Propenoyl chloride] 814-68-6 1 Allyl alcohol [2-Propen-1-ol] 107-18-6 1 Allylamine [2-Propen-1-amine] 107-11-9 1 Aluminum (powder) 7429-90-5 ACG Ammonia (anhydrous) 7664-41-7 1 Ammonia (conc.