TBC1D24 Mutations in a Sibship with Multifocal Polymyoclonus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Clinicians Using the Classification Will Identify a Seizure As Focal Or Generalized Onset If There Is About an 80% Confidence Level About the Type of Onset
GENERALIZED ONSET SEIZURES Generalized onset seizures are not characterized by level of awareness, because awareness is almost always impaired. Generalized tonic-clonic: Immediate loss of Generalized epileptic spasms: Brief seizures with awareness, with stiffening of all limbs (tonic phase), flexion at the trunk and flexion or extension of the followed by sustained rhythmic jerking of limbs and limbs. Video-EEG recording may be required to face (clonic phase). Duration is typically 1 to 3 minutes. determine focal versus generalized onset. The seizure may produce a cry at the start, falling, tongue biting, and incontinence. Generalized typical absence: Sudden onset when activity stops with a brief pause and staring, Generalized clonic: Rhythmical sustained jerking of sometimes with eye fluttering and head nodding or limbs and/or head with no tonic stiffening phase. other automatic behaviors. If it lasts for more than These seizures most often occur in young children. several seconds, awareness and memory are impaired. Recovery is immediate. The EEG during these seizures Generalized tonic: Stiffening of all limbs, without always shows generalized spike-waves. clonic jerking. Generalized atypical absence: Like typical absence Generalized myoclonic: Irregular, unsustained jerking seizures, but may have slower onset and recovery and of limbs, face, eyes, or eyelids. The jerking of more pronounced changes in tone. Atypical absence generalized myoclonus may not always be left-right seizures can be difficult to distinguish from focal synchronous, but it occurs on both sides. impaired awareness seizures, but absence seizures usually recover more quickly and the EEG patterns are Generalized myoclonic-tonic-clonic: This seizure is like different. -
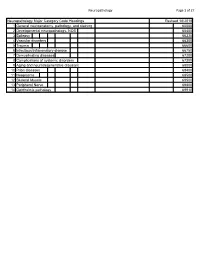
Neuropathology Category Code List
Neuropathology Page 1 of 27 Neuropathology Major Category Code Headings Revised 10/2018 1 General neuroanatomy, pathology, and staining 65000 2 Developmental neuropathology, NOS 65400 3 Epilepsy 66230 4 Vascular disorders 66300 5 Trauma 66600 6 Infectious/inflammatory disease 66750 7 Demyelinating diseases 67200 8 Complications of systemic disorders 67300 9 Aging and neurodegenerative diseases 68000 10 Prion diseases 68400 11 Neoplasms 68500 12 Skeletal Muscle 69500 13 Peripheral Nerve 69800 14 Ophthalmic pathology 69910 Neuropathology Page 2 of 27 Neuropathology 1 General neuroanatomy, pathology, and staining 65000 A Neuroanatomy, NOS 65010 1 Neocortex 65011 2 White matter 65012 3 Entorhinal cortex/hippocampus 65013 4 Deep (basal) nuclei 65014 5 Brain stem 65015 6 Cerebellum 65016 7 Spinal cord 65017 8 Pituitary 65018 9 Pineal 65019 10 Tracts 65020 11 Vascular supply 65021 12 Notochord 65022 B Cell types 65030 1 Neurons 65031 2 Astrocytes 65032 3 Oligodendroglia 65033 4 Ependyma 65034 5 Microglia and mononuclear cells 65035 6 Choroid plexus 65036 7 Meninges 65037 8 Blood vessels 65038 C Cerebrospinal fluid 65045 D Pathologic responses in neurons and axons 65050 1 Axonal degeneration/spheroid/reaction 65051 2 Central chromatolysis 65052 3 Tract degeneration 65053 4 Swollen/ballooned neurons 65054 5 Trans-synaptic neuronal degeneration 65055 6 Olivary hypertrophy 65056 7 Acute ischemic (hypoxic) cell change 65057 8 Apoptosis 65058 9 Protein aggregation 65059 10 Protein degradation/ubiquitin pathway 65060 E Neuronal nuclear inclusions 65100 -

Myoclonic Status Epilepticus in Juvenile Myoclonic Epilepsy
Original article Epileptic Disord 2009; 11 (4): 309-14 Myoclonic status epilepticus in juvenile myoclonic epilepsy Julia Larch, Iris Unterberger, Gerhard Bauer, Johannes Reichsoellner, Giorgi Kuchukhidze, Eugen Trinka Department of Neurology, Medical University of Innsbruck, Austria Received April 9, 2009; Accepted November 18, 2009 ABSTRACT – Background. Myoclonic status epilepticus (MSE) is rarely found in juvenile myoclonic epilepsy (JME) and its clinical features are not well described. We aimed to analyze MSE incidence, precipitating factors and clini- cal course by studying patients with JME from a large outpatient epilepsy clinic. Methods. We retrospectively screened all patients with JME treated at the Department of Neurology, Medical University of Innsbruck, Austria between 1970 and 2007 for a history of MSE. We analyzed age, sex, age at seizure onset, seizure types, EEG, MRI/CT findings and response to antiepileptic drugs. Results. Seven patients (five women, two men; median age at time of MSE 31 years; range 17-73) with MSE out of a total of 247 patients with JME were identi- fied. The median follow-up time was seven years (range 0-35), the incidence was 3.2/1,000 patient years. Median duration of epilepsy before MSE was 26 years (range 10-58). We identified three subtypes: 1) MSE with myoclonic seizures only in two patients, 2) MSE with generalized tonic clonic seizures in three, and 3) generalized tonic clonic seizures with myoclonic absence status in two patients. All patients responded promptly to benzodiazepines. One patient had repeated episodes of MSE. Precipitating events were identified in all but one patient. Drug withdrawal was identified in four patients, one of whom had additional sleep deprivation and alcohol intake. -

Neuronal Hyperexcitability in a Mouse Model of SCN8A Epileptic Encephalopathy
Neuronal hyperexcitability in a mouse model of SCN8A epileptic encephalopathy Luis F. Lopez-Santiagoa,1, Yukun Yuana,1, Jacy L. Wagnonb, Jacob M. Hulla, Chad R. Frasiera, Heather A. O’Malleya, Miriam H. Meislerb,c, and Lori L. Isoma,c,d,2 aDepartment of Pharmacology, University of Michigan, Ann Arbor, MI 48109; bDepartment of Human Genetics, University of Michigan, Ann Arbor, MI 48109; cDepartment of Neurology, University of Michigan, Ann Arbor, MI 48109; and dDepartment of Molecular and Integrative Physiology, University of Michigan, Ann Arbor, MI 48109 Edited by William A. Catterall, University of Washington School of Medicine, Seattle, WA, and approved January 17, 2017 (received for review October 12, 2016) Patients with early infantile epileptic encephalopathy (EIEE) experi- include seizure onset between birth and 18 mo of age, mild to se- ence severe seizures and cognitive impairment and are at increased vere cognitive and developmental delay, and mild to severe risk for sudden unexpected death in epilepsy (SUDEP). EIEE13 [Online movement disorders (11, 13). Approximately 50% of patients are Mendelian Inheritance in Man (OMIM) # 614558] is caused by de nonambulatory and 12% of published cases (5/43) experienced novo missense mutations in the voltage-gated sodium channel gene SUDEP during childhood or adolescence (10, 13, 16, 17). SCN8A. Here, we investigated the neuronal phenotype of a mouse Almost all of the identified mutations in SCN8A are missense model expressing the gain-of-function SCN8A patient mutation, mutations. Ten of these mutations have been tested functionally in p.Asn1768Asp (Nav1.6-N1768D). Our results revealed regional transfected cells, and eight were found to introduce changes in the and neuronal subtype specificity in the effects of the N1768D muta- biophysical properties of Na 1.6 that are predicted to result in Scn8aN1768D/+ v tion. -

Reviews Familial Cortical Myoclonic Tremor and Epilepsy, an Enigmatic Disorder: from Phenotypes to Pathophysiology and Genetics
Freely available online Reviews Familial Cortical Myoclonic Tremor and Epilepsy, an Enigmatic Disorder: From Phenotypes to Pathophysiology and Genetics. A Systematic Review 1 1 2,3 1* Tom van den Ende , Sarvi Sharifi , Sandra M. A. van der Salm & Anne-Fleur van Rootselaar 1 Department of Neurology and Clinical Neurophysiology, Amsterdam Neuroscience, Academic Medical Center, Amsterdam, The Netherlands, 2 Brain Center Rudolf Magnus, Department of Neurology and Neurosurgery, University Medical Center, Utrecht, The Netherlands, 3 Stichting Epilepsie Instellingen Nederland (SEIN), Zwolle, The Netherlands Abstract Background: Autosomal dominant familial cortical myoclonic tremor and epilepsy (FCMTE) is characterized by distal tremulous myoclonus, generalized seizures, and signs of cortical reflex myoclonus. FCMTE has been described in over 100 pedigrees worldwide, under several different names and acronyms. Pathological changes have been located in the cerebellum. This systematic review discusses the clinical spectrum, treatment, pathophysiology, and genetic findings. Methods: We carried out a PubMed search, using a combination of the following search terms: cortical tremor, myoclonus, epilepsy, benign course, adult onset, familial, and autosomal dominant; this resulted in a total of 77 studies (761 patients; 126 pedigrees) fulfilling the inclusion and exclusion criteria. Results: Phenotypic differences across pedigrees exist, possibly related to underlying genetic differences. A ‘‘benign’’ phenotype has been described in several Japanese families and pedigrees linked to 8q (FCMTE1). French patients (5p linkage; FCMTE3) exhibit more severe progression, and in Japanese/Chinese pedigrees (with unknown linkage) anticipation has been suggested. Preferred treatment is with valproate (mind teratogenicity), levetiracetam, and/or clonazepam. Several genes have been identified, which differ in potential pathogenicity. Discussion: Based on the core features (above), the syndrome can be considered a distinct clinical entity. -

EFNS Guidelines for the Use of Intravenous Immunoglobulin In
European Journal of Neurology 2008, 15: 893–908 doi:10.1111/j.1468-1331.2008.02246.x EFNS TASK FORCE/CME ARTICLE EFNS guidelines for the use of intravenous immunoglobulin in treatment of neurological diseases EFNS task force on the use of intravenous immunoglobulin in treatment of neurological diseases Members of the Task Force: I. Elovaaraa, S. Apostolskib, P. van Doornc, N. E. Gilhusd, A. Hietaharjue, J. Honkaniemif, I. N. van Schaikg, N. Scoldingh, P. Soelberg Sørenseni and B. Uddj aDepartment of Neurology and Rehabilitation, Tampere University Hospital and Medical School, University of Tampere, Tampere, Finland; bInstitute of Neurology, School of Medicine, University of Belgrade, Belgrade, Serbia; cDepartment of Neurology, Erasmus Medical Centre, Rotterdam, The Netherlands; dDepartment of Neurology, Haukeland University Hospital, Bergen, Norway; eDepartment of Neurology and Rehabilitation, Tampere University Hospital, Tampere, Finland; fDepartment of Neurology, Vaasa Central Hospital, Vaasa, Finland; gDepartment of Neurology, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands; hUniversity of Bristol Institute Of Clinical Neuroscience, Frenchary Hospital UK Bristol, UK; iDepartment of Neurology, National University Hospital, Rigshospitalet, Copenhagen, Denmark; and jDepartment of Neurology and Rehabilitation, Tampere University Hospital and Medical School, University of Tampere, Tampere, Finland Keywords: Despite high-dose intravenous immunoglobulin (IVIG) is widely used in treatment of acute disseminated a -

Epileptic Encephalopathy
Epileptic Encephalopathy ศ. นพ. อนันตน์ ิตย ์ วิสทุ ธิพนั ธ์ Division of Neurology, Department of Pediatrics, Faculty of Medicine-Ramathibodi Hospital, Mahidol University, Bangkok, Thailand Scope of Talk • Overview of Epileptic Encephalopathy • Issues in epileptic encephalopathy • Frequently seen epileptic encephalopathy • Application to daily practice in reality • Q & A What is Epileptic Encephalopathy? Epileptic Encephalopathy • 1840: First described by West WJ from the letter to The Lancet describing “West syndrome” • 1955: Illingworth RS reported 12 cases of “sudden mental deterioration with convulsion in infancy” • 1966: Gastaut H “EE related to the concept that the underlying epileptic activity may contribute to the neurodevelopmental compromise noted in children with early onset, severe epilepsy and abundant spike and wave activities” Epileptic Encephalopathy • 1841 West syndrome • 1957 Landau-Kleffner syndrome • 1960 Lennox-Gastaut syndrome • 1987 Myoclonic-atonic (-astatic) epilepsy • 1971 Electrical Status Epilepticus during sleep (ESES) & Continuous Spikes and Waves during Sleep (CSWS) • 1976 Early Infantile Epileptic Encephalopathy (EIEE) • 1978 Severe Myoclonic Epilepsy in Infancy (SMEI) or Dravet syndrome • ………………………………………….. Epileptic Encephalopathy ILAE 2001: “A condition where the epileptiform abnormalities themselves are believed to contribute to the progressive disturbance of cerebral function” Single Insult Epileptic Encephalopathy or Cause Epilepsy Cognitive dysfunction Delayed Behavioral Development impairment Epileptic -

Juvenile Myoclonic Epilepsy: Under-Appreciated and Under-Diagnosed R Renganathan, N Delanty
78 Postgrad Med J: first published as 10.1136/pmj.79.928.78 on 1 February 2003. Downloaded from REVIEW Juvenile myoclonic epilepsy: under-appreciated and under-diagnosed R Renganathan, N Delanty ............................................................................................................................. Postgrad Med J 2003;79:78–80 Juvenile myoclonic epilepsy (JME) is a hereditary, AETIOPATHOGENESIS idiopathic, generalised epilepsy and is found in JME is an inherited disorder, but the exact mode of inheritance is not clear. There is a positive fam- 5%–11% of patients with epilepsy. It is characterised by ily history in 50% of cases or less. One study has myoclonic jerks, occasional generalised tonic-clonic confirmed a causative role of EJM1 in the patho- seizures, and sometimes absence seizures. JME genesis of idiopathic generalised seizures in the majority of German families of JME patients. This continues to be under-appreciated and study has refined a candidate region of 10.1 cM in under-diagnosed. Accurate diagnosis is important as it the chromosomal region 6p21 between the flank- usually responds well to treatment with appropriate ing loci HLA-DQ and D6s1019.3 However, some studies have revealed controversial results and anticonvulsants and misdiagnosis often results in genetic heterogeneity is suspected. Linkage to unnecessary morbidity. In addition lifelong therapy is chromosome 15 has at most a minor role in usually indicated as the natural history is one of relapse JME.4 Few studies have focused on pathological find- -

VIDEO ABSTRACTS Epilepsy, Dyskinesia and ASD in an Infant
VIDEO ABSTRACTS Epilepsy, dyskinesia and ASD in an infant with probably ALG1 mutation (CDG-Ik) – a case presentation Mihaela Adela Vintan 1, Camelia Al-Khzouz 1, Diana Miclea 1 University of Medicine and Pharmacy - Cluj Napoca (Romania) Introduction: Epilepsy associated with ASD is a group of highly heterogeneous diseases with various phenotypes. Their genotype has recently started to be studied. SCN1A and MECP2 are responsible for most of the phenotypes. Voltage-gated ion channel activity genes played an important role in epilepsy with ASD patients, including SCN1A, SCN2A, CACNA1A, CACNA1H, CACNA1D, and KCNQ2. Still, in 50% of these children, the etiology remains unknown, yet. Methods: We present a boy, from a non-consanguineous family, a healthy older brother; with un- eventful pregnancy and birth. He had normal development in the first 3 months of life. From the fourth month, he started to present seizures, with focal onset, tonic, with impaired awareness and epileptic spasms, rare but associated in time, with microcephaly and cognitive and motor deterioration with autistic features. Hepatic cytolysis was associated from early infancy. Around the age of 1 year he developed dyskinesia. Seizures became resistant to therapy and the development was slow, both motor and cognitive. Results: The etiology was investigated. MRI was normal. Congenital infections were excluded (TORCH). Due to the fact that there were signs pointing on a metabolic disorder, aminoacidopathies and organic acidurias, acylcarnitine profile, lysosomal disorders, very long chain fatty acids, copper metabolism were evaluated and revealed normal values. MLPA (SALSA MLPA KIT P245 Microdeletion syndromes, MRC-Holland) was performed and showed negative results. -

Generalized Epilepsy?
A GUIDE FOR PATIENTS & FAMILIES WHAT IS IDIOPATHIC GENERALIZED EPILEPSY? Idiopathic generalized epilepsy, abbreviated IGE, is a group of epilepsy that has very distinct features. It is also called “primary” generalized epilepsy. The term “idiopathic” is misleading, because it usually means “cause unknown,” which is not true here. They are genetic, and in fact were recently renamed “genetic generalized epilepsy.” Defining Idiopathic generalized epilepsy Doctors of USF Health People with IGE have normal intelligence, normal CHARACTERISTICS neurological examination, and normal MRIs. These epilepsies have the following characteristics: Although they are clearly genetic, they are not transmitted predictably like, for example, hemophil- They are genetic and not caused by any brain ia or cystic fibrosis. physical abnormality. This means that the brain is anatomically normal. TYPE OF SEIZURES They basically represent a genetic low threshold (or high susceptibility) for seizures. Patients with IGE have one or more of 3 types of (primary generalized) seizures: myoclonic, ab- There is often, but not always, a family history of sence and generalized tonic-clonic seizures. epilepsy. (If everyone had 50 siblings, patients with IGE would more often have a family member af- One type may be the only or main type in a given fected). patient. For some specific types (for example, juvenile my- Generalized tonic-clonic (“grand-mal”) seizures are oclonic epilepsy) the chromosome and gene have convulsions of the whole body lasting 1-2 minutes, even been identified. Others will almost certainly and are the most common and most dramatic type be identified in the near future. of seizures. Seizures are especially sensitive to sleep depri- Absence seizures are brief staring spells with arrest vation of activity, often with eye fluttering, which last just a few seconds. -

Clinical and Genetic Overview of Paroxysmal Movement Disorders and Episodic Ataxias
International Journal of Molecular Sciences Review Clinical and Genetic Overview of Paroxysmal Movement Disorders and Episodic Ataxias Giacomo Garone 1,2 , Alessandro Capuano 2 , Lorena Travaglini 3,4 , Federica Graziola 2,5 , Fabrizia Stregapede 4,6, Ginevra Zanni 3,4, Federico Vigevano 7, Enrico Bertini 3,4 and Francesco Nicita 3,4,* 1 University Hospital Pediatric Department, IRCCS Bambino Gesù Children’s Hospital, University of Rome Tor Vergata, 00165 Rome, Italy; [email protected] 2 Movement Disorders Clinic, Neurology Unit, Department of Neuroscience and Neurorehabilitation, IRCCS Bambino Gesù Children’s Hospital, 00146 Rome, Italy; [email protected] (A.C.); [email protected] (F.G.) 3 Unit of Neuromuscular and Neurodegenerative Diseases, Department of Neuroscience and Neurorehabilitation, IRCCS Bambino Gesù Children’s Hospital, 00146 Rome, Italy; [email protected] (L.T.); [email protected] (G.Z.); [email protected] (E.B.) 4 Laboratory of Molecular Medicine, IRCCS Bambino Gesù Children’s Hospital, 00146 Rome, Italy; [email protected] 5 Department of Neuroscience, University of Rome Tor Vergata, 00133 Rome, Italy 6 Department of Sciences, University of Roma Tre, 00146 Rome, Italy 7 Neurology Unit, Department of Neuroscience and Neurorehabilitation, IRCCS Bambino Gesù Children’s Hospital, 00165 Rome, Italy; [email protected] * Correspondence: [email protected]; Tel.: +0039-06-68592105 Received: 30 April 2020; Accepted: 13 May 2020; Published: 20 May 2020 Abstract: Paroxysmal movement disorders (PMDs) are rare neurological diseases typically manifesting with intermittent attacks of abnormal involuntary movements. Two main categories of PMDs are recognized based on the phenomenology: Paroxysmal dyskinesias (PxDs) are characterized by transient episodes hyperkinetic movement disorders, while attacks of cerebellar dysfunction are the hallmark of episodic ataxias (EAs). -

A Type of Progressive Myoclonic Epilepsy, Lafora Disease: a Case Report
Eastern Journal of Medicine 18 (2013) 34-36 Case Report A type of progressive myoclonic epilepsy, Lafora disease: A case report Ömer Bektaşa,*, Arzu Yılmaza, Aylin Heper Okcuc, Serap Teberb, Erhan Aksoya, Gülhis Dedaa aDepartment of Pediatric Neurology, School of Medicine, Ankara University,Turkey bDepartment of Pediatric Neurology, Dıskapı Hematology-Oncology Hospital, Turkey cDepartment of Pediatric Pathology, School of Medicine, Ankara University, Turkey Abstract. Lafora disease is a rare group of progressive myoclonic epilepsies characterized with progressive neurological dysfunction, myoclonus, focal and generalized seizures. Generally, a generalized tonic clonic seizure is the first symptom of the disease. An 11-year-old male patient had been followed-up at another center for epilepsy for 8 years. The patient had a history of myoclonic seizures for nearly every day for the last 2 years and cognitive detoriation for the last 8 months. He admitted to our hospital with the desire of his family. Eccrine sweat gland biopsy was performed. The biopsy of the sweat gland was positive for PAS and contained diastase resistant polyglican content (Lafora bodies), and thus, a diagnosis of Lafora disease was established. The patient presented here constitutes a rare case of pediatric epilepsy, which caused neurodegeneration in late-childhood and onset with typical epilepsy symptoms. This report also aimed to show that biopsy obtained from proper area is important for diagnosis Our patient developed cognitive dysfunction a short period of eight months. To our knowledge, this is the shortest period in literature. Key words: Lafora Disease, progressive myoclonic epilepsy, neurodegeneration 1. Introduction 2. Case report Progressive myoclonic epilepsy is a rare group An 11-year-old male patient had been followed- of diseases characterized with progressive up for epilepsy for 8 years.