Transformation Activation to Oncogenic Growth Critical for The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Clock Is Involved in Predictive Circadian Adjustment of Renal Function
Molecular clock is involved in predictive circadian adjustment of renal function Annie Mercier Zubera,1,2, Gabriel Centenoa,2, Sylvain Pradervandb, Svetlana Nikolaevaa,c, Lionel Maquelina, Le´ onard Cardinauxa, Olivier Bonnya,d, and Dmitri Firsova,3 aDepartment of Pharmacology and Toxicology, University of Lausanne, 1005 Lausanne, Switzerland; bDNA Array Facility, University of Lausanne, 1015 Lausanne, Switzerland; cInstitute of Evolutionary Physiology and Biochemistry, 194223 St-Petersburg, Russia; and dService of Nephrology, Lausanne University Hospital, 1005 Lausanne, Switzerland Edited by Maurice B. Burg, National Heart, Lung, and Blood Institute, Bethesda, MD, and approved August 6, 2009 (received for review May 4, 2009) Renal excretion of water and major electrolytes exhibits a significant anticipate upcoming circadian environmental challenges (activity, circadian rhythm. This functional periodicity is believed to result, at feeding, etc). least in part, from circadian changes in secretion/reabsorption capac- The most obvious manifestation of circadian rhythmicity of renal ities of the distal nephron and collecting ducts. Here, we studied the function is a well-marked difference in the volume of urine forma- molecular mechanisms underlying circadian rhythms in the distal tion/excretion between the day and the night. The urinary excretion nephron segments, i.e., distal convoluted tubule (DCT) and connect- ϩ ϩ Ϫ Ϫ 2ϩ 2ϩ of all major solutes (Na ,K ,Cl , urea, PO4 ,Ca ,Mg )also ing tubule (CNT) and the cortical collecting duct (CCD). Temporal follows a circadian oscillating pattern. Although, renal excretion expression analysis performed on microdissected mouse DCT/CNT or rhythms are apparently synchronized with circadian rhythms of CCD revealed a marked circadian rhythmicity in the expression of a activity/feeding, they have been shown to persist over long periods large number of genes crucially involved in various homeostatic of time under experimental conditions in which external factors functions of the kidney. -

The Microtubule-Associated Protein MAPRE2 Is Involved in Perineural Invasion of Pancreatic Cancer Cells
1111-1116.qxd 8/9/2009 08:18 Ì ™ÂÏ›‰·1111 INTERNATIONAL JOURNAL OF ONCOLOGY 35: 1111-1116, 2009 The microtubule-associated protein MAPRE2 is involved in perineural invasion of pancreatic cancer cells IVANE ABIATARI1*, SONJA GILLEN1*, TIAGO DeOLIVEIRA1, THERESA KLOSE1, KONG BO1, NATHALIA A. GIESE2, HELMUT FRIESS1 and JÖRG KLEEFF1,3 1Department of General Surgery, Technische Universität München, Munich; 2Department of General Surgery, University of Heidelberg, Heidelberg, Germany; 3Center of Cancer Systems Biology, Department of Medicine, Caritas St. Elizabeth's Medical Center, Tufts University School of Medicine, Boston, MA 02135-2997, USA Received May 4, 2009; Accepted July 10, 2009 DOI: 10.3892/ijo_00000426 Abstract. Perineural invasion of tumor cells is a characteristic Defining molecular mechanisms that allow pancreatic cancer feature of human pancreatic cancer. Unrevealing the molecular cells to grow along and into nerves is therefore important for mechanisms that enable cancer cells to invade and grow along the development of novel therapeutic strategies in pancreatic nerves is important for the development of novel therapeutic cancer, a disease whose incidence virtually mirrors its mortality strategies in this disease. We have previously identified tran- rate (5). We have previously identified and described in detail scriptional changes in highly nerve invasive pancreatic cancer the transcriptome signature of perineural invasion in pancreatic cells. Here we further analyzed one of the identified de- cancer by generating highly nerve invasive pancreatic cancer regulated genes, MAPRE2, a microtubule-associated protein. cells (6). One of the genes that displayed a significant up- MAPRE2 expression was significantly increased in high versus regulation in pancreatic cancer cells with a high potential for less nerve invasive pancreatic cancer cells, and changes of nerve invasion was MAPRE2. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -

Supplementary Table 1: List of the 316 Genes Regulated During Hyperglycemic Euinsulinemic Clamp in Skeletal Muscle
Supplementary Table 1: List of the 316 genes regulated during hyperglycemic euinsulinemic clamp in skeletal muscle. UGCluster Name Symbol Fold Change Cytoband Response to stress Hs.517581 Heme oxygenase (decycling) 1 HMOX1 3.80 22q12 Hs.374950 Metallothionein 1X MT1X 2.20 16q13 Hs.460867 Metallothionein 1B (functional) MT1B 1.70 16q13 Hs.148778 Oxidation resistance 1 OXR1 1.60 8q23 Hs.513626 Metallothionein 1F (functional) MT1F 1.47 16q13 Hs.534330 Metallothionein 2A MT2A 1.45 16q13 Hs.438462 Metallothionein 1H MT1H 1.42 16q13 Hs.523836 Glutathione S-transferase pi GSTP1 -1.74 11q13 Hs.459952 Stannin SNN -1.92 16p13 Immune response, cytokines & related Hs.478275 TNF (ligand) superfamily, member 10 (TRAIL) TNFSF10 1.58 3q26 Hs.278573 CD59 antigen p18-20 (protectin) CD59 1.49 11p13 Hs.534847 Complement component 4B, telomeric C4A 1.47 6p21.3 Hs.535668 Immunoglobulin lambda variable 6-57 IGLV6-57 1.40 22q11.2 Hs.529846 Calcium modulating ligand CAMLG -1.40 5q23 Hs.193516 B-cell CLL/lymphoma 10 BCL10 -1.40 1p22 Hs.840 Indoleamine-pyrrole 2,3 dioxygenase INDO -1.40 8p12-p11 Hs.201083 Mal, T-cell differentiation protein 2 MAL2 -1.44 Hs.522805 CD99 antigen-like 2 CD99L2 -1.45 Xq28 Hs.50002 Chemokine (C-C motif) ligand 19 CCL19 -1.45 9p13 Hs.350268 Interferon regulatory factor 2 binding protein 2 IRF2BP2 -1.47 1q42.3 Hs.567249 Contactin 1 CNTN1 -1.47 12q11-q12 Hs.132807 MHC class I mRNA fragment 3.8-1 3.8-1 -1.48 6p21.3 Hs.416925 Carcinoembryonic antigen-related cell adhesion molecule 19 CEACAM19 -1.49 19q13.31 Hs.89546 Selectin E (endothelial -
Activation of NF-Κb Signaling Promotes Prostate Cancer Progression in the Mouse and Predicts Poor Progression and Death in Pati
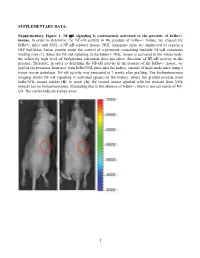
SUPPLEMENTARY DATA: Supplementary Figure 1. NF-B signaling is continuously activated in the prostate of - mouse. In order to determine the NF- '- mouse, we crossed the '- mice with NGL, a NF-B reporter mouse. NGL transgenic mice are engineered to express a GFP/luciferase fusion protein under the control of a promoter containing multiple NF-B consensus binding sites (1). Since the NF-'- NGL mouse is activated in the whole body, the relatively high level of background activation does not allow detection of NF- prostate. Therefore, in order to determine the NF-the '- mouse, we grafted the prostates from 'o the kidney capsule of male nude mice using a tissue rescue technique. NF-B activity was measured at 7 weeks after grafting. The bioluminescence imaging shows NF-B signaling is activated (green) in the kidney, where the grafted prostate from ' use resides (B). In panel (A), the control mouse (grafted with the prostate from NGL mouse) has no bioluminescence, illustrating that in the absence of '-, there is not activation of NF- B. The circles indicate kidney areas. 1 Supplementary Figure 2. NF-B signaling activated in the prostate of Myc/IB bigenic mouse. The prostates from Myc alone (Myc) and bigenic (Myc/IB) mice were harvested at 6 months of age. Activation of NF-B signaling in the prostate was determined by IHC staining of p65-pho antibody. 2 Supplementary Figure 3. Continuous activation of NF-B signaling promotes PCa progression in the Hi-Myc transgenic mouse. The prostates from Myc alone (Myc) and bigeneic (Myc/IB) mice were harvested at 6 months of age. -

The Kinesin Spindle Protein Inhibitor Filanesib Enhances the Activity of Pomalidomide and Dexamethasone in Multiple Myeloma
Plasma Cell Disorders SUPPLEMENTARY APPENDIX The kinesin spindle protein inhibitor filanesib enhances the activity of pomalidomide and dexamethasone in multiple myeloma Susana Hernández-García, 1 Laura San-Segundo, 1 Lorena González-Méndez, 1 Luis A. Corchete, 1 Irena Misiewicz- Krzeminska, 1,2 Montserrat Martín-Sánchez, 1 Ana-Alicia López-Iglesias, 1 Esperanza Macarena Algarín, 1 Pedro Mogollón, 1 Andrea Díaz-Tejedor, 1 Teresa Paíno, 1 Brian Tunquist, 3 María-Victoria Mateos, 1 Norma C Gutiérrez, 1 Elena Díaz- Rodriguez, 1 Mercedes Garayoa 1* and Enrique M Ocio 1* 1Centro Investigación del Cáncer-IBMCC (CSIC-USAL) and Hospital Universitario-IBSAL, Salamanca, Spain; 2National Medicines Insti - tute, Warsaw, Poland and 3Array BioPharma, Boulder, Colorado, USA *MG and EMO contributed equally to this work ©2017 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2017.168666 Received: March 13, 2017. Accepted: August 29, 2017. Pre-published: August 31, 2017. Correspondence: [email protected] MATERIAL AND METHODS Reagents and drugs. Filanesib (F) was provided by Array BioPharma Inc. (Boulder, CO, USA). Thalidomide (T), lenalidomide (L) and pomalidomide (P) were purchased from Selleckchem (Houston, TX, USA), dexamethasone (D) from Sigma-Aldrich (St Louis, MO, USA) and bortezomib from LC Laboratories (Woburn, MA, USA). Generic chemicals were acquired from Sigma Chemical Co., Roche Biochemicals (Mannheim, Germany), Merck & Co., Inc. (Darmstadt, Germany). MM cell lines, patient samples and cultures. Origin, authentication and in vitro growth conditions of human MM cell lines have already been characterized (17, 18). The study of drug activity in the presence of IL-6, IGF-1 or in co-culture with primary bone marrow mesenchymal stromal cells (BMSCs) or the human mesenchymal stromal cell line (hMSC–TERT) was performed as described previously (19, 20). -

The Microtubule End-Binding Protein EB2 Is a Central Regulator Of
4000 Research Article The microtubule end-binding protein EB2 is a central regulator of microtubule reorganisation in apico-basal epithelial differentiation Deborah A. Goldspink1, Jonathan R. Gadsby1, Gemma Bellett1, Jennifer Keynton1, Benjamin J. Tyrrell1, Elizabeth K. Lund1, Penny P. Powell2, Paul Thomas1 and Mette M. Mogensen1,* 1School of Biological Sciences, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK 2Norwich Medical School, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK *Author for correspondence ([email protected]) Accepted 31 May 2013 Journal of Cell Science 126, 4000–4014 ß 2013. Published by The Company of Biologists Ltd doi: 10.1242/jcs.129759 Summary Microtubule end-binding (EB) proteins influence microtubule dynamic instability, a process that is essential for microtubule reorganisation during apico-basal epithelial differentiation. Here, we establish for the first time that expression of EB2, but not that of EB1, is crucial for initial microtubule reorganisation during apico-basal epithelial differentiation, and that EB2 downregulation promotes bundle formation. EB2 siRNA knockdown during early stages of apico-basal differentiation prevented microtubule reorganisation, whereas its downregulation at later stages promoted microtubule stability and bundle formation. Interestingly, although EB1 is not essential for microtubule reorganisation, its knockdown prevented apico-basal bundle formation and epithelial elongation. siRNA depletion of EB2 in undifferentiated epithelial cells induced the formation of straight, less dynamic microtubules with EB1 and ACF7 lattice association and co-alignment with actin filaments, a phenotype that could be rescued by inhibition with formin. Importantly, in situ inner ear and intestinal crypt epithelial tissue revealed direct correlations between a low level of EB2 expression and the presence of apico-basal microtubule bundles, which were absent where EB2 was elevated. -

Supplementary Table 1: Genes Located on Chromosome 18P11-18Q23, an Area Significantly Linked to TMPRSS2-ERG Fusion
Supplementary Table 1: Genes located on Chromosome 18p11-18q23, an area significantly linked to TMPRSS2-ERG fusion Symbol Cytoband Description LOC260334 18p11 HSA18p11 beta-tubulin 4Q pseudogene IL9RP4 18p11.3 interleukin 9 receptor pseudogene 4 LOC100132166 18p11.32 hypothetical LOC100132166 similar to Rho-associated protein kinase 1 (Rho- associated, coiled-coil-containing protein kinase 1) (p160 LOC727758 18p11.32 ROCK-1) (p160ROCK) (NY-REN-35 antigen) ubiquitin specific peptidase 14 (tRNA-guanine USP14 18p11.32 transglycosylase) THOC1 18p11.32 THO complex 1 COLEC12 18pter-p11.3 collectin sub-family member 12 CETN1 18p11.32 centrin, EF-hand protein, 1 CLUL1 18p11.32 clusterin-like 1 (retinal) C18orf56 18p11.32 chromosome 18 open reading frame 56 TYMS 18p11.32 thymidylate synthetase ENOSF1 18p11.32 enolase superfamily member 1 YES1 18p11.31-p11.21 v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 LOC645053 18p11.32 similar to BolA-like protein 2 isoform a similar to 26S proteasome non-ATPase regulatory LOC441806 18p11.32 subunit 8 (26S proteasome regulatory subunit S14) (p31) ADCYAP1 18p11 adenylate cyclase activating polypeptide 1 (pituitary) LOC100130247 18p11.32 similar to cytochrome c oxidase subunit VIc LOC100129774 18p11.32 hypothetical LOC100129774 LOC100128360 18p11.32 hypothetical LOC100128360 METTL4 18p11.32 methyltransferase like 4 LOC100128926 18p11.32 hypothetical LOC100128926 NDC80 homolog, kinetochore complex component (S. NDC80 18p11.32 cerevisiae) LOC100130608 18p11.32 hypothetical LOC100130608 structural maintenance -

EB2 (MAPRE2) (NM 014268) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC200259 EB2 (MAPRE2) (NM_014268) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: EB2 (MAPRE2) (NM_014268) Human Tagged ORF Clone Tag: Myc-DDK Symbol: MAPRE2 Synonyms: CSCSC2; EB1; EB2; RP1 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC200259 ORF sequence Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGCCTGGGCCGACCCAAACCCTGTCCCCAAATGGCGAGAACAACAACGACATCATCCAGGATAATAACG GGACCATCATTCCTTTCCGGAAGCACACAGTGCGCGGGGAGCGTTCCTACAGTTGGGGAATGGCGGTCAA TGTGTATTCTACCTCGATAACCCAAGAGACTATGAGCAGACATGACATCATTGCATGGGTTAATGACATA GTATCTTTAAACTACACAAAAGTGGAACAGCTTTGTTCAGGAGCGGCCTATTGCCAATTCATGGACATGC TCTTCCCTGGCTGCATTAGTTTGAAGAAAGTAAAATTTCAAGCAAAGCTGGAACATGAATATATTCACAA TTTTAAACTTCTGCAAGCATCATTTAAGCGAATGAACGTTGATAAGGTAATTCCAGTGGAGAAGCTAGTG AAAGGACGTTTCCAGGACAACCTGGATTTTATTCAATGGTTTAAGAAATTCTATGATGCTAACTACGATG GGAAGGAGTATGATCCTGTAGAGGCACGACAAGGGCAAGATGCAATTCCTCCTCCTGACCCTGGTGAACA GATCTTCAACCTGCCAAAAAAGTCTCACCATGCAAACTCCCCCACAGCAGGTGCAGCTAAATCAAGTCCA GCAGCTAAACCAGGATCCACACCTTCTCGACCCTCATCAGCCAAAAGGGCTTCTTCCAGTGGCTCAGCAT CCAAATCCGATAAAGATTTAGAAACGCAGGTCATACAGCTTAATGAACAGGTACATTCATTAAAACTTGC CCTTGAAGGCGTGGAAAAGGAAAGGGATTTCTACTTTGGGAAGTTGAGAGAGATCGAGCTACTCTGCCAA GAACACGGGCAGGAAAATGATGACCTCGTGCAGAGACTAATGGACATCCTGTATGCTTCAGAAGAACACG -

Identification of Endogenous Adenomatous Polyposis Coli Interaction Partners and Β-Catenin−Independent Targets by Proteomics
Published OnlineFirst June 3, 2019; DOI: 10.1158/1541-7786.MCR-18-1154 Cancer Genes and Networks Molecular Cancer Research Identification of Endogenous Adenomatous Polyposis Coli Interaction Partners and b-Catenin– Independent Targets by Proteomics Olesja Popow1,2,3,Joao~ A. Paulo2, Michael H. Tatham4, Melanie S. Volk3, Alejandro Rojas-Fernandez5, Nicolas Loyer3, Ian P. Newton3, Jens Januschke3, Kevin M. Haigis1,6, and Inke Nathke€ 3 Abstract Adenomatous Polyposis Coli (APC) is the most frequently tion between endogenous MINK1 and APC and further mutated gene in colorectal cancer. APC negatively regulates confirmed the negative, and b-catenin–independent, regu- the Wnt signaling pathway by promoting the degradation of lation of MINK1 by APC. Increased Mink1/Msn levels were b-catenin, but the extent to which APC exerts Wnt/b-cate- also observed in mouse intestinal tissue and Drosophila nin–independent tumor-suppressive activity is unclear. To follicular cells expressing mutant Apc/APC when compared identify interaction partners and b-catenin–independent with wild-type tissue/cells. Collectively, our results highlight targets of endogenous, full-length APC, we applied label- the extent and importance of Wnt-independent APC func- free and multiplexed tandem mass tag-based mass spec- tions in epithelial biology and disease. trometry. Affinity enrichment-mass spectrometry identified more than 150 previously unidentified APC interaction Implications: The tumor-suppressive function of APC, the partners. Moreover, our global proteomic analysis revealed most frequently mutated gene in colorectal cancer, is mainly that roughly half of the protein expression changes that attributed to its role in b-catenin/Wnt signaling. Our study occur in response to APC loss are independent of b-catenin. -
![EB2 (MAPRE2) Mouse Monoclonal Antibody [Clone ID: OTI5F3] – TA502961 | Origene](https://docslib.b-cdn.net/cover/0549/eb2-mapre2-mouse-monoclonal-antibody-clone-id-oti5f3-ta502961-origene-3220549.webp)
EB2 (MAPRE2) Mouse Monoclonal Antibody [Clone ID: OTI5F3] – TA502961 | Origene
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA502961 EB2 (MAPRE2) Mouse Monoclonal Antibody [Clone ID: OTI5F3] Product data: Product Type: Primary Antibodies Clone Name: OTI5F3 Applications: FC, IF, IHC, WB Recommended Dilution: WB 1:2000, IHC 1:150, IF 1:100, FLOW 1:100 Reactivity: Human Host: Mouse Isotype: IgG2a Clonality: Monoclonal Immunogen: Full length human recombinant protein of human MAPRE2 (NP_055083) produced in HEK293T cell. Formulation: PBS (PH 7.3) containing 1% BSA, 50% glycerol and 0.02% sodium azide. Concentration: 1 mg/ml Purification: Purified from mouse ascites fluids or tissue culture supernatant by affinity chromatography (protein A/G) Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 36.9 kDa Gene Name: microtubule associated protein RP/EB family member 2 Database Link: NP_055083 Entrez Gene 10982 Human Q15555 Background: The protein encoded by this gene shares significant homology to the adenomatous polyposis coli (APC) protein-binding EB1 gene family. This protein is a microtubule-associated protein that is necessary for spindle symmetry during mitosis. It is thought to play a role in the tumorigenesis of colorectal cancers and the proliferative control of normal cells. Alternative splicing of this gene results in multiple transcript variants. [provided by RefSeq, Jan 2012] This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 5 EB2 (MAPRE2) Mouse Monoclonal Antibody [Clone ID: OTI5F3] – TA502961 Synonyms: EB1; EB2; RP1 Protein Families: Druggable Genome Product images: HEK293T cells were transfected with the pCMV6- ENTRY control (Left lane) or pCMV6-ENTRY MAPRE2 ([RC200259], Right lane) cDNA for 48 hrs and lysed.