The RNA-Binding Profile of Acinus, a Peripheral Component of the Exon Junction Complex, Reveals Its Role in Splicing Regulation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

DFFA Monoclonal Antibody (M05), Degradation of the Chromosomal DNA Into Nucleosomal Clone 3A11 Units
DFFA monoclonal antibody (M05), degradation of the chromosomal DNA into nucleosomal clone 3A11 units. DNA fragmentation factor (DFF) is a heterodimeric protein of 40-kD (DFFB) and 45-kD (DFFA) subunits. Catalog Number: H00001676-M05 DFFA is the substrate for caspase-3 and triggers DNA fragmentation during apoptosis. DFF becomes activated Regulatory Status: For research use only (RUO) when DFFA is cleaved by caspase-3. The cleaved fragments of DFFA dissociate from DFFB, the active Product Description: Mouse monoclonal antibody component of DFF. DFFB has been found to trigger both raised against a partial recombinant DFFA. DNA fragmentation and chromatin condensation during apoptosis. Two alternatively spliced transcript variants Clone Name: 3A11 encoding distinct isoforms have been found for this gene. [provided by RefSeq] Immunogen: DFFA (NP_004392.1, 231 a.a. ~ 331 a.a) partial recombinant protein with GST tag. MW of the GST tag alone is 26 KDa. Sequence: TSSDVALASHILTALREKQAPELSLSSQDLELVTKEDPK ALAVALNWDIKKTETVQEACERELALRLQQTQSLHSLR SISASKASPPGDLQNPKRARQDPT Host: Mouse Reactivity: Human Applications: ELISA, PLA-Ce, S-ELISA, WB-Re, WB-Tr (See our web site product page for detailed applications information) Protocols: See our web site at http://www.abnova.com/support/protocols.asp or product page for detailed protocols Isotype: IgG2a Kappa Storage Buffer: In 1x PBS, pH 7.4 Storage Instruction: Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. Entrez GeneID: 1676 Gene Symbol: DFFA Gene Alias: DFF-45, DFF1, ICAD Gene Summary: Apoptosis is a cell death process that removes toxic and/or useless cells during mammalian development. The apoptotic process is accompanied by shrinkage and fragmentation of the cells and nuclei and Page 1/1 Powered by TCPDF (www.tcpdf.org). -

Whole Exome Sequencing in ADHD Trios from Single and Multi-Incident Families Implicates New Candidate Genes and Highlights Polygenic Transmission
European Journal of Human Genetics (2020) 28:1098–1110 https://doi.org/10.1038/s41431-020-0619-7 ARTICLE Whole exome sequencing in ADHD trios from single and multi-incident families implicates new candidate genes and highlights polygenic transmission 1,2 1 1,2 1,3 1 Bashayer R. Al-Mubarak ● Aisha Omar ● Batoul Baz ● Basma Al-Abdulaziz ● Amna I. Magrashi ● 1,2 2 2,4 2,4,5 6 Eman Al-Yemni ● Amjad Jabaan ● Dorota Monies ● Mohamed Abouelhoda ● Dejene Abebe ● 7 1,2 Mohammad Ghaziuddin ● Nada A. Al-Tassan Received: 26 September 2019 / Revised: 26 February 2020 / Accepted: 10 March 2020 / Published online: 1 April 2020 © The Author(s) 2020. This article is published with open access Abstract Several types of genetic alterations occurring at numerous loci have been described in attention deficit hyperactivity disorder (ADHD). However, the role of rare single nucleotide variants (SNVs) remains under investigated. Here, we sought to identify rare SNVs with predicted deleterious effect that may contribute to ADHD risk. We chose to study ADHD families (including multi-incident) from a population with a high rate of consanguinity in which genetic risk factors tend to 1234567890();,: 1234567890();,: accumulate and therefore increasing the chance of detecting risk alleles. We employed whole exome sequencing (WES) to interrogate the entire coding region of 16 trios with ADHD. We also performed enrichment analysis on our final list of genes to identify the overrepresented biological processes. A total of 32 rare variants with predicted damaging effect were identified in 31 genes. At least two variants were detected per proband, most of which were not exclusive to the affected individuals. -

DFFB (NM 004402) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC208266 DFFB (NM_004402) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: DFFB (NM_004402) Human Tagged ORF Clone Tag: Myc-DDK Symbol: DFFB Synonyms: CAD; CPAN; DFF-40; DFF2; DFF40 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC208266 ORF sequence Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGCTCCAGAAGCCCAAGAGCGTGAAGCTGCGGGCCCTGCGCAGCCCGAGGAAGTTCGGCGTGGCTGGCC GGAGCTGCCAGGAGGTGCTGCGCAAGGGCTGTCTCCGCTTCCAGCTCCCTGAGCGCGGTTCCCGGCTGTG CCTGTACGAGGATGGCACGGAGCTGACGGAAGATTACTTCCCCAGTGTTCCCGACAACGCCGAGCTGGTG CTGCTCACCTTGGGCCAGGCCTGGCAGGGCTATGTGAGCGACATCAGGCGCTTCCTCAGTGCATTTCACG AGCCACAGGTGGGGCTCATCCAGGCCGCCCAGCAGCTGCTGTGTGATGAGCAGGCCCCACAGAGGCAGAG GCTGCTGGCTGACCTCCTGCACAACGTCAGCCAGAACATCGCGGCCGAGACCCGGGCTGAGGACCCGCCG TGGTTTGAAGGCTTGGAGTCCCGATTTCAGAGCAAGTCTGGCTATCTGAGATACAGCTGTGAGAGCCGGA TCCGGAGTTACCTGAGGGAGGTGAGCTCCTACCCCTCCACAGTGGGTGCGGAGGCTCAGGAGGAATTCCT GCGGGTCCTCGGCTCCATGTGCCAGAGGCTCCGGTCCATGCAGTACAATGGCAGCTACTTCGACAGAGGA GCCAAGGGCGGCAGCCGCCTCTGCACACCGGAAGGCTGGTTCTCCTGCCAGGGTCCCTTTGACATGGACA GCTGCTTATCAAGACACTCCATCAACCCCTACAGTAACAGGGAGAGCAGGATCCTCTTCAGCACCTGGAA CCTGGATCACATAATAGAAAAGAAACGCACCATCATTCCTACACTGGTGGAAGCAATTAAGGAACAAGAT GGAAGAGAAGTGGACTGGGAGTATTTTTATGGCCTGCTTTTTACCTCAGAGAACCTAAAACTAGTGCACA -
NUTM1 Is a Recurrent Fusion Gene Partner in B-Cell Precursor Acute
LETTERS TO THE EDITOR However, 20-25% of BCP-ALL patients do not have one NUTM1 is a recurrent fusion gene partner in B-cell of these sentinel cytogenetic aberrations and are there- precursor acute lymphoblastic leukemia associated fore said to have B-other ALL. This B-other ALL subgroup with increased expression of genes on chromosome has an intermediate risk of relapse, but includes both band 10p12.31-12.2 high- and low-risk subgroups that are currently being identified. Our laboratory identified a subtype with a For 20-25% of patients with pediatric B-cell precursor similar expression profile and prognosis as BCR-ABL1, acute lymphoblastic leukemia (BCP-ALL), the driving namely BCR-ABL1-like, within the B-other ALL sub- cytogenetic aberration is unknown. Identification of the group.2 The B-other ALL subgroup also includes other primary lesion could provide better risk stratification and rare cytogenetic subtypes, such as intrachromosomal even identify possible treatment options. We therefore amplification of chromosome 21 and a dicentric chromo- aimed to find novel recurrent genetic aberrations in BCP- 1 ALL cases. We identified an in-frame SLC12A6-NUTM1 some (9;20). It is important to identify more primary fusion, resulting in expression of 3’ exons of NUTM1, lesions in the remaining B-other ALL for better risk strat- and six additional NUTM1-rearranged fusion cases. ification and identification of possible treatment options. These NUTM1-rearranged cases were associated with In this study, we aimed to identify recurrent fusions in high expression of a cluster of genes on chromosome BCP-ALL cases without currently known lesions through band 10p12.31-12.2, including the BMI1 gene. -

Viewed and Published Immediately Upon Acceptance Cited in Pubmed and Archived on Pubmed Central Yours — You Keep the Copyright
BMC Genomics BioMed Central Research article Open Access Differential gene expression in ADAM10 and mutant ADAM10 transgenic mice Claudia Prinzen1, Dietrich Trümbach2, Wolfgang Wurst2, Kristina Endres1, Rolf Postina1 and Falk Fahrenholz*1 Address: 1Johannes Gutenberg-University, Institute of Biochemistry, Mainz, Johann-Joachim-Becherweg 30, 55128 Mainz, Germany and 2Helmholtz Zentrum München – German Research Center for Environmental Health, Institute for Developmental Genetics, Ingolstädter Landstraße 1, 85764 Neuherberg, Germany Email: Claudia Prinzen - [email protected]; Dietrich Trümbach - [email protected]; Wolfgang Wurst - [email protected]; Kristina Endres - [email protected]; Rolf Postina - [email protected]; Falk Fahrenholz* - [email protected] * Corresponding author Published: 5 February 2009 Received: 19 June 2008 Accepted: 5 February 2009 BMC Genomics 2009, 10:66 doi:10.1186/1471-2164-10-66 This article is available from: http://www.biomedcentral.com/1471-2164/10/66 © 2009 Prinzen et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: In a transgenic mouse model of Alzheimer disease (AD), cleavage of the amyloid precursor protein (APP) by the α-secretase ADAM10 prevented amyloid plaque formation, and alleviated cognitive deficits. Furthermore, ADAM10 overexpression increased the cortical synaptogenesis. These results suggest that upregulation of ADAM10 in the brain has beneficial effects on AD pathology. Results: To assess the influence of ADAM10 on the gene expression profile in the brain, we performed a microarray analysis using RNA isolated from brains of five months old mice overexpressing either the α-secretase ADAM10, or a dominant-negative mutant (dn) of this enzyme. -

ICAD Antibody Cat
ICAD Antibody Cat. No.: 2003 ICAD Antibody Immunofluorescence of ICAD in mouse kidney tissue with ICAD antibody at 5 μg/ml. Immunohistochemistry of ICAD in mouse kidney tissue with ICAD antibody at 5 μg/ml. Green: ICAD antibody (2003) Red: Phylloidin staining Blue: DAPI staining Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Mouse ICAD antibody was raised against a 20 amino acid peptide near the carboxy terminus of mouse ICAD. IMMUNOGEN: The immunogen is located within the last 50 amino acids of ICAD. TESTED APPLICATIONS: ELISA, IF, IHC-P, WB September 28, 2021 1 https://www.prosci-inc.com/icad-antibody-2003.html ICAD antibody can be used for detection of of ICAD by Western blot at 1:1000 dilution. A 45 kDa band can be detected. Antibody can also be used for immunohistochemistry starting at 5 μg/mL. For immunofluorescence start at 5 μg/mL. APPLICATIONS: Antibody validated: Western Blot in mouse samples; Immunohistochemistry in mouse samples and Immunofluorescence in mouse samples. All other applications and species not yet tested. POSITIVE CONTROL: 1) Cat. No. 1402 - Mouse Lung Tissue Lysate PREDICTED MOLECULAR 45 kDa WEIGHT: Properties PURIFICATION: ICAD Antibody is affinity chromatography purified via peptide column. CLONALITY: Polyclonal ISOTYPE: IgG CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: ICAD Antibody is supplied in PBS containing 0.02% sodium azide. CONCENTRATION: 1 mg/ml ICAD antibody can be stored at 4˚C for three months and -20˚C, stable for up to one year. STORAGE CONDITIONS: As with all antibodies care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. -

Proteomic Analysis of Ubiquitin Ligase KEAP1 Reveals Associated Proteins That Inhibit NRF2 Ubiquitination
Published OnlineFirst February 4, 2013; DOI: 10.1158/0008-5472.CAN-12-4400 Cancer Molecular and Cellular Pathobiology Research Proteomic Analysis of Ubiquitin Ligase KEAP1 Reveals Associated Proteins That Inhibit NRF2 Ubiquitination Bridgid E. Hast1, Dennis Goldfarb2, Kathleen M. Mulvaney1, Michael A. Hast4, Priscila F. Siesser1, Feng Yan1, D. Neil Hayes3, and Michael B. Major1,2 Abstract Somatic mutations in the KEAP1 ubiquitin ligase or its substrate NRF2 (NFE2L2) commonly occur in human cancer, resulting in constitutive NRF2-mediated transcription of cytoprotective genes. However, many tumors display high NRF2 activity in the absence of mutation, supporting the hypothesis that alternative mechanisms of pathway activation exist. Previously, we and others discovered that via a competitive binding mechanism, the proteins WTX (AMER1), PALB2, and SQSTM1 bind KEAP1 to activate NRF2. Proteomic analysis of the KEAP1 protein interaction network revealed a significant enrichment of associated proteins containing an ETGE amino acid motif, which matches the KEAP1 interaction motif found in NRF2. Like WTX, PALB2, and SQSTM1, we found that the dipeptidyl peptidase 3 (DPP3) protein binds KEAP1 via an "ETGE" motif to displace NRF2, thus inhibiting NRF2 ubiquitination and driving NRF2-dependent transcription. Comparing the spectrum of KEAP1-interacting proteins with the genomic profile of 178 squamous cell lung carcinomas characterized by The Cancer Genome Atlas revealed amplification and mRNA overexpression of the DPP3 gene in tumors with high NRF2 activity but lacking NRF2 stabilizing mutations. We further show that tumor-derived mutations in KEAP1 are hypomorphic with respect to NRF2 inhibition and that DPP3 overexpression in the presence of these mutants further promotes NRF2 activation. -

Produktinformation
Produktinformation Diagnostik & molekulare Diagnostik Laborgeräte & Service Zellkultur & Verbrauchsmaterial Forschungsprodukte & Biochemikalien Weitere Information auf den folgenden Seiten! See the following pages for more information! Lieferung & Zahlungsart Lieferung: frei Haus Bestellung auf Rechnung SZABO-SCANDIC Lieferung: € 10,- HandelsgmbH & Co KG Erstbestellung Vorauskassa Quellenstraße 110, A-1100 Wien T. +43(0)1 489 3961-0 Zuschläge F. +43(0)1 489 3961-7 [email protected] • Mindermengenzuschlag www.szabo-scandic.com • Trockeneiszuschlag • Gefahrgutzuschlag linkedin.com/company/szaboscandic • Expressversand facebook.com/szaboscandic DFFA Recombinant Protein (OPCD02721) Data Sheet Product Number OPCD02721 Product Page http://www.avivasysbio.com/dffa-recombinant-protein-opcd02721.html Product Name DFFA Recombinant Protein (OPCD02721) Size 10 ug Gene Symbol DFFA Alias Symbols A330085O09Rik, DFF35, Dff45, DFF-45, DNA fragmentation factor 45 kDa subunit, DNA fragmentation factor subunit alpha, Icad, ICAD, ICAD-L, IC AD-S, Inhibitor of CAD Molecular Weight 31 kDa Product Format Lyophilized Tag N-terminal His Tag Conjugation Unconjugated NCBI Gene Id 13347 Host E.coli Purity > 95% Source Prokaryotic Expressed Recombinant Official Gene Full Name DNA fragmentation factor, alpha subunit Description of Target Inhibitor of the caspase-activated DNase (DFF40). Reconstitution and Storage Reconstitute in PBS or others. Store at 2-8C for one month. Aliquot and store at -80C for 12 months. Avoid repeated freeze/thaw cycles. Additional Information Endotoxin Level: < 1.0 EU per 1 ug (determined by the LAL method) Additional Information Residues: Lys19 - Ser262 Lead Time Domestic: within 1-2 weeks delivery International: 1-3 weeks Formulation Lyophilized in PBS, pH7.4, containing 0.01% SKL, 1mM DTT, 5% Trehalose and Proclin300. -

Molecular Mechanisms in Muscular Dystrophy : a Gene Expression Profiling Study Turk, R
Molecular mechanisms in muscular dystrophy : a gene expression profiling study Turk, R. Citation Turk, R. (2006, September 27). Molecular mechanisms in muscular dystrophy : a gene expression profiling study. Retrieved from https://hdl.handle.net/1887/4577 Version: Corrected Publisher’s Version Licence agreement concerning inclusion of doctoral License: thesis in the Institutional Repository of the University of Leiden Downloaded from: https://hdl.handle.net/1887/4577 Note: To cite this publication please use the final published version (if applicable). Molecular Mechanisms In Muscular Dystrophy A Gene Expression Profiling Study Molecular Mechanisms In Muscular Dystrophy A Gene Expression Profiling Study Proefschrift ter verkrijging van de graad van Doctor aan de Universiteit Leiden, op gezag van de Rector Magnificus Dr. D.D.Breimer, hoogleraar in de faculteit der Wiskunde en Natuurwetenschappen en die der Geneeskunde, volgens besluit van het College voor Promoties te verdedigen op woensdag 27 september 2006 klokke 15.00 uur door Rolf Turk Geboren te Leiden in 1975 Promotiecommissie Promotor Prof. Dr. G.J.B. van Ommen Co-promotores Dr. J.T. den Dunnen Dr. P.A.C. ‘t Hoen Referent Prof. Dr. R.M.W. Hofstra (Rijksuniversiteit Groningen) Overige leden Prof. Dr. M. Koenig (Université Louis Pasteur de Strasbourg) An experiment is a question which science poses to Nature, and a measurement is the recording of Nature’s answer. Max Planck Aan Maaike, Gerard en Annet Printed by: Drukkerij Duineveld ISBN-10: 90-9021042-3 ISBN-13: 978-90-9021042-1 Turk, Rolf Molecular mechanisms in muscular dystrophy. A gene expression profiling study. Thesis, Leiden University Medical Center September 27, 2006 © Rolf Turk No part of this thesis may be reproduced or transmitted in any form or by any means, without the written permission of the copyright owner Molecular Mechanisms In Muscular Dystrophies Preface 9 Chapter 1 Introduction 11 1. -

Anti-ICAD Antibody (ARG54401)
Product datasheet [email protected] ARG54401 Package: 50 μg anti-ICAD antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes ICAD Tested Reactivity Hu Tested Application ICC/IF, WB Specificity This antibody recognizes non-cleaved (45kDa) and cleaved DFF45. Host Rabbit Clonality Polyclonal Isotype IgG Target Name ICAD Antigen Species Human Immunogen Peptide corresponding to aa 313-331 at the C-terminus of human DFF45 (accession no. NP_004392). Conjugation Un-conjugated Alternate Names DFF-45; DNA fragmentation factor 45 kDa subunit; Inhibitor of CAD; ICAD; DFF1; DNA fragmentation factor subunit alpha Application Instructions Application table Application Dilution ICC/IF Assay-dependent WB Assay-dependent Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control HeLa, Jurkat, A431 and K562 Calculated Mw 37 kDa Properties Form Liquid Purification Immunoaffinity chroma-tography Buffer PBS (pH 7.4) and 0.02% Sodium azide Preservative 0.02% Sodium azide Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C or below. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. www.arigobio.com 1/2 Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Database links GeneID: 1676 Human Swiss-port # O00273 Human Gene Symbol DFFA Gene Full Name DNA fragmentation factor, 45kDa, alpha polypeptide Background A human DNA fragmentation factor (DFF) which is cleaved by caspase-3 during apoptosis was identified recently. -

DFFA Antibody Cat
DFFA Antibody Cat. No.: 27-308 DFFA Antibody Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human Antibody produced in rabbits immunized with a synthetic peptide corresponding a region IMMUNOGEN: of human DFFA. TESTED APPLICATIONS: ELISA, WB DFFA antibody can be used for detection of DFFA by ELISA at 1:312500. DFFA antibody can APPLICATIONS: be used for detection of DFFA by western blot at 5.0 μg/mL, and HRP conjugated secondary antibody should be diluted 1:50,000 - 100,000. POSITIVE CONTROL: 1) Cat. No. 1205 - Jurkat Cell Lysate PREDICTED MOLECULAR 37 kDa, 29 kDa WEIGHT: Properties PURIFICATION: Antibody is purified by protein A chromatography method. CLONALITY: Polyclonal CONJUGATE: Unconjugated PHYSICAL STATE: Liquid September 29, 2021 1 https://www.prosci-inc.com/dffa-antibody-27-308.html Purified antibody supplied in 1x PBS buffer with 0.09% (w/v) sodium azide and 2% BUFFER: sucrose. CONCENTRATION: batch dependent For short periods of storage (days) store at 4˚C. For longer periods of storage, store DFFA STORAGE CONDITIONS: antibody at -20˚C. As with any antibody avoid repeat freeze-thaw cycles. Additional Info OFFICIAL SYMBOL: DFFA ALTERNATE NAMES: DFFA, DFF-45, DFF1, Ion ChannelAD, ICAD ACCESSION NO.: NP_004392 PROTEIN GI NO.: 4758148 GENE ID: 1676 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References Apoptosis is a cell death process that removes toxic and/or useless cells during mammalian development. The apoptotic process is accompanied by shrinkage and fragmentation of the cells and nuclei and degradation of the chromosomal DNA into nucleosomal units. DNA fragmentation factor (DFF) is a heterodimeric protein of 40-kD (DFFB) and 45-kD (DFFA) subunits. -
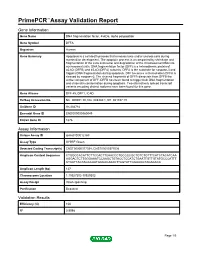
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name DNA fragmentation factor, 45kDa, alpha polypeptide Gene Symbol DFFA Organism Human Gene Summary Apoptosis is a cell death process that removes toxic and/or useless cells during mammalian development. The apoptotic process is accompanied by shrinkage and fragmentation of the cells and nuclei and degradation of the chromosomal DNA into nucleosomal units. DNA fragmentation factor (DFF) is a heterodimeric protein of 40-kD (DFFB) and 45-kD (DFFA) subunits. DFFA is the substrate for caspase-3 and triggers DNA fragmentation during apoptosis. DFF becomes activated when DFFA is cleaved by caspase-3. The cleaved fragments of DFFA dissociate from DFFB the active component of DFF. DFFB has been found to trigger both DNA fragmentation and chromatin condensation during apoptosis. Two alternatively spliced transcript variants encoding distinct isoforms have been found for this gene. Gene Aliases DFF-45, DFF1, ICAD RefSeq Accession No. NC_000001.10, NG_008340.1, NT_021937.19 UniGene ID Hs.484782 Ensembl Gene ID ENSG00000160049 Entrez Gene ID 1676 Assay Information Unique Assay ID qHsaCID0012769 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000377038, ENST00000377036 Amplicon Context Sequence CTGGCCACATTCTTCCACTTCAACCCTGCCCCGCTGTCTGTTTCATCTACATCAA AGGACTCTTGGGAAATCCAAGCTGTACCTCCATCTGAATTGTTGTATGCCCATTT CTCATTACTAGCCAATGCCACAAACTTAGTATTGGAAGGTAGACACA Amplicon Length (bp) 127 Chromosome Location 1:10527302-10529302 Assay Design Intron-spanning Purification Desalted Validation Results