Targeting Fibroblast CD248 Attenuates CCL17-Expressing Macrophages
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Multimerin-2 Is a Ligand for Group 14 Family C-Type Lectins CLEC14A, CD93 and CD248 Spanning the Endothelial Pericyte Interface
OPEN Oncogene (2017) 36, 6097–6108 www.nature.com/onc ORIGINAL ARTICLE Multimerin-2 is a ligand for group 14 family C-type lectins CLEC14A, CD93 and CD248 spanning the endothelial pericyte interface KA Khan1, AJ Naylor2, A Khan1, PJ Noy1, M Mambretti1, P Lodhia1, J Athwal1, A Korzystka1, CD Buckley2, BE Willcox3, F Mohammed3 and R Bicknell1 TheC-typelectindomaincontaininggroup14familymembers CLEC14A and CD93 are proteins expressed by endothelium and are implicated in tumour angiogenesis. CD248 (alternatively known as endosialin or tumour endothelial marker-1) is also a member of this family and is expressed by tumour-associated fibroblasts and pericytes. Multimerin-2 (MMRN2) is a unique endothelial specific extracellular matrix protein that has been implicated in angiogenesis and tumour progression. We show that the group 14 C-type lectins CLEC14A, CD93 and CD248 directly bind to MMRN2 and only thrombomodulin of the family does not. Binding to MMRN2 is dependent on a predicted long-loop region in the C-type lectin domainandisabrogatedbymutationwithinthedomain. CLEC14A and CD93 bind to the same non-glycosylated coiled-coil region of MMRN2, but the binding of CD248 occurs on a distinct non-competing region. CLEC14A and CD248 can bind MMRN2 simultaneously and this occurs at the interface between endothelium and pericytes in human pancreatic cancer. A recombinant peptide of MMRN2 spanning the CLEC14A and CD93 binding region blocks CLEC14A extracellular domain binding to the endothelial cellsurfaceaswellasincreasingadherenceofhumanumbilical vein endothelial cells to the active peptide. This MMRN2 peptide is anti-angiogenic in vitro and reduces tumour growth in mouse models. These findings identify novel protein interactions involving CLEC14A, CD93 and CD248 with MMRN2 as targetable components of vessel formation. -

CD Markers Are Routinely Used for the Immunophenotyping of Cells
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called CD markers are routinely used for the immunophenotyping of cells. Despite this use, they are not limited to roles in the immune system and perform a variety of roles in cell differentiation, adhesion, migration, blood clotting, gamete fertilization, amino acid transport and apoptosis, among many others. As such, Proteintech’s mini catalog featuring its antibodies targeting CD markers is applicable to a wide range of research disciplines. PRODUCT FOCUS PECAM1 Platelet endothelial cell adhesion of blood vessels – making up a large portion molecule-1 (PECAM1), also known as cluster of its intracellular junctions. PECAM-1 is also CD Number of differentiation 31 (CD31), is a member of present on the surface of hematopoietic the immunoglobulin gene superfamily of cell cells and immune cells including platelets, CD31 adhesion molecules. It is highly expressed monocytes, neutrophils, natural killer cells, on the surface of the endothelium – the thin megakaryocytes and some types of T-cell. Catalog Number layer of endothelial cells lining the interior 11256-1-AP Type Rabbit Polyclonal Applications ELISA, FC, IF, IHC, IP, WB 16 Publications Immunohistochemical of paraffin-embedded Figure 1: Immunofluorescence staining human hepatocirrhosis using PECAM1, CD31 of PECAM1 (11256-1-AP), Alexa 488 goat antibody (11265-1-AP) at a dilution of 1:50 anti-rabbit (green), and smooth muscle KD/KO Validated (40x objective). alpha-actin (red), courtesy of Nicola Smart. PECAM1: Customer Testimonial Nicola Smart, a cardiovascular researcher “As you can see [the immunostaining] is and a group leader at the University of extremely clean and specific [and] displays Oxford, has said of the PECAM1 antibody strong intercellular junction expression, (11265-1-AP) that it “worked beautifully as expected for a cell adhesion molecule.” on every occasion I’ve tried it.” Proteintech thanks Dr. -

HOD1164 Biorad Mouse Poster A0 V2.Indd
Biomarker Expression Patterns in Mouse Immune Cells Non Non Non Cells Cells Cells T Cells T Cells T Cells T B Cells B Cells B Cells B NK Cells Cells NK Cells NK Cells NK Mast Cells Mast Cells Mast Cells Mast and Innate and Innate and Innate Stem Cells, Cells, Stem Cells, Stem Cells, Stem Progenitors Progenitors Progenitors Myeloid and Myeloid and Myeloid and Hematopoietic Hematopoietic Hematopoietic Hematopoietic Erythroid Cells Erythroid Cells Erythroid Cells Lymphoid Cells Lymphoid Cells Lymphoid Cells Lymphoid Precursors and and Precursors and Precursors and Precursors Megakaryocyte Megakaryocyte Megakaryocyte Presenting Cells Presenting Cells Presenting Cells Specialist Antigen Antigen Specialist Antigen Specialist Antigen Specialist Markers Markers Markers Langerhans cells Langerhans cells Langerhans cells Langerhans LT-HSC GMP CMP CLP MEP MPP cells B cells B Pro/Pre dendritic cells Myeloid dendriticPlasmacytoid cells Eosinophil Neutrophil Basophil Macrophage Monocyte Granulocyte cellsMast cells NK cells NK Precursor lymphoid progenitor Innate lymphoid cells Innate cells T Double negative thymocyte Double positive thymocyte Gamma/Delta cells T NKT cells Megakaryocyte Platelets Proerythroblast Erythrocyte Endothelial cells Epithelial cells LT-HSC GMP CMP CLP MEP MPP cells B cells B Pro/Pre dendritic cells Myeloid dendriticPlasmacytoid cells Eosinophil Neutrophil Basophil Macrophage Monocyte Granulocyte cellsMast cells NK cells NK Precursor lymphoid progenitor Innate lymphoid cells Innate cells T Double negative thymocyte Double positive -

Is CD248 Involved in the Resolution of Inflammation During Development Of
Is CD248 involved in the resolution of inflammation during development of lung sarcoidosis? and The role of adipose tissue-derived stromal cells in the survival of lymphocytes Presented by Bonita H. R. Apta College of Medical and Dental Sciences University of Birmingham 13th August 2012 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. TABLE OF CONTENTS PROJECT 1: ..........................................................................................................................1 1. INTRODUCTION............................................................................................................1 1.1 IMMUNOPATHOLOGY OF SARCOIDOSIS......................................................................1 1.1.1 Aetiology of sarcoidosis..................................................................................2 1.1.2 Pathology of sarcoidosis.................................................................................2 1.1.3 Pulmonary fibrosis in the sarcoid lung............................................................3 -

Mouse CD Marker Chart Bdbiosciences.Com/Cdmarkers
BD Mouse CD Marker Chart bdbiosciences.com/cdmarkers 23-12400-01 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD1d CD1.1, CD1.2, Ly-38 Lipid, Glycolipid Ag + + + + + + + + CD104 Integrin b4 Laminin, Plectin + DNAX accessory molecule 1 (DNAM-1), Platelet and T cell CD226 activation antigen 1 (PTA-1), T lineage-specific activation antigen 1 CD112, CD155, LFA-1 + + + + + – + – – CD2 LFA-2, Ly-37, Ly37 CD48, CD58, CD59, CD15 + + + + + CD105 Endoglin TGF-b + + antigen (TLiSA1) Mucin 1 (MUC1, MUC-1), DF3 antigen, H23 antigen, PUM, PEM, CD227 CD54, CD169, Selectins; Grb2, β-Catenin, GSK-3β CD3g CD3g, CD3 g chain, T3g TCR complex + CD106 VCAM-1 VLA-4 + + EMA, Tumor-associated mucin, Episialin + + + + + + Melanotransferrin (MT, MTF1), p97 Melanoma antigen CD3d CD3d, CD3 d chain, T3d TCR complex + CD107a LAMP-1 Collagen, Laminin, Fibronectin + + + CD228 Iron, Plasminogen, pro-UPA (p97, MAP97), Mfi2, gp95 + + CD3e CD3e, CD3 e chain, CD3, T3e TCR complex + + CD107b LAMP-2, LGP-96, LAMP-B + + Lymphocyte antigen 9 (Ly9), -

Multi-Omic Single Cell Analysis Resolves Novel Stromal Cell
www.nature.com/scientificreports OPEN Multi‑omic single cell analysis resolves novel stromal cell populations in healthy and diseased human tendon Adrian R. Kendal1,2*, Thomas Layton1, Hussein Al‑Mossawi1, Louise Appleton1, Stephanie Dakin1, Rick Brown2, Constantinos Loizou2, Mark Rogers2, Robert Sharp2 & Andrew Carr1 Tendinopathy accounts for over 30% of primary care consultations and represents a growing healthcare challenge in an active and increasingly ageing population. Recognising critical cells involved in tendinopathy is essential in developing therapeutics to meet this challenge. Tendon cells are heterogenous and sparsely distributed in a dense collagen matrix; limiting previous methods to investigate cell characteristics ex vivo. We applied next generation CITE‑sequencing; combining surface proteomics with in‑depth, unbiased gene expression analysis of > 6400 single cells ex vivo from 11 chronically tendinopathic and 8 healthy human tendons. Immunohistochemistry validated the single cell fndings. For the frst time we show that human tendon harbours at least fve distinct COL1A1/2 expressing tenocyte populations in addition to endothelial cells, T‑cells, and monocytes. These consist of KRT7/SCX+ cells expressing microfbril associated genes, PTX3+ cells co‑expressing high levels of pro‑infammatory markers, APOD+ fbro–adipogenic progenitors, TPPP3/PRG4+ chondrogenic cells, and ITGA7+ smooth muscle‑mesenchymal cells. Surface proteomic analysis identifed markers by which these sub‑classes could be isolated and targeted in future. Chronic tendinopathy was associated with increased expression of pro‑infammatory markers PTX3, CXCL1, CXCL6, CXCL8, and PDPN by microfbril associated tenocytes. Diseased endothelium had increased expression of chemokine and alarmin genes including IL33. Musculoskeletal disorders are responsible for the second largest number of years lived with disability worldwide1. -
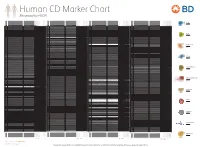
Reviewed by HLDA1
Human CD Marker Chart Reviewed by HLDA1 T Cell Key Markers CD3 CD4 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD8 CD1a R4, T6, Leu6, HTA1 b-2-Microglobulin, CD74 + + + – + – – – CD74 DHLAG, HLADG, Ia-g, li, invariant chain HLA-DR, CD44 + + + + + + CD158g KIR2DS5 + + CD248 TEM1, Endosialin, CD164L1, MGC119478, MGC119479 Collagen I/IV Fibronectin + ST6GAL1, MGC48859, SIAT1, ST6GALL, ST6N, ST6 b-Galactosamide a-2,6-sialyl- CD1b R1, T6m Leu6 b-2-Microglobulin + + + – + – – – CD75 CD22 CD158h KIR2DS1, p50.1 HLA-C + + CD249 APA, gp160, EAP, ENPEP + + tranferase, Sialo-masked lactosamine, Carbohydrate of a2,6 sialyltransferase + – – + – – + – – CD1c M241, R7, T6, Leu6, BDCA1 b-2-Microglobulin + + + – + – – – CD75S a2,6 Sialylated lactosamine CD22 (proposed) + + – – + + – + + + CD158i KIR2DS4, p50.3 HLA-C + – + CD252 TNFSF4, -

The Constitutive Extracellular Protein Release by Acute Myeloid
cancers Article The Constitutive Extracellular Protein Release by Acute Myeloid Leukemia Cells—A Proteomic Study of Patient Heterogeneity and Its Modulation by Mesenchymal Stromal Cells Elise Aasebø 1,2 , Annette K. Brenner 1, Even Birkeland 2, Tor Henrik Anderson Tvedt 3, Frode Selheim 2, Frode S. Berven 2 and Øystein Bruserud 2,3,* 1 Department of Clinical Science, University of Bergen, 5021 Bergen, Norway; [email protected] (E.A.); [email protected] (A.K.B.) 2 The Proteomics Facility of the University of Bergen (PROBE), University of Bergen, 5009 Bergen, Norway; [email protected] (E.B.); [email protected] (F.S.); [email protected] (F.S.B.) 3 Department of Medicine, Haukeland University Hospital, 5021 Bergen, Norway; [email protected] * Correspondence: [email protected] or [email protected] Simple Summary: The formation of normal blood cells in the bone marrow is supported by a network of non-hematopoietic cells including connective tissue cells, blood vessel cells and bone-forming cells. These cell types support and regulate the growth of acute myeloid leukemia (AML) cells Citation: Aasebø, E.; Brenner, A.K.; and communicate with leukemic cells through the release of proteins to their common extracellular Birkeland, E.; Tvedt, T.H.A.; Selheim, F.; Berven, F.S.; Bruserud, Ø. The microenvironment. One of the AML-supporting normal cell types is a subset of connective tissue Constitutive Extracellular Protein cells called mesenchymal stem cells. In the present study, we observed that AML cells release a wide Release by Acute Myeloid Leukemia range of diverse proteins into their microenvironment, but patients differ both with regard to the Cells—A Proteomic Study of Patient number and amount of released proteins. -

Photothermal Modulation of Human Stem Cells Using Light-Responsive 2D Nanomaterials
Photothermal modulation of human stem cells using light-responsive 2D nanomaterials James K. Carrowa,1, Kanwar Abhay Singha,1, Manish K. Jaiswala, Adelina Ramireza, Giriraj Lokhandea, Alvin T. Yeha, Tapasree Roy Sarkarb, Irtisha Singha,c,2, and Akhilesh K. Gaharwara,d,e,2 aBiomedical Engineering, College of Engineering, Texas A&M University, College Station, TX 77843; bDepartment of Biology, Texas A&M University, College Station, TX 77843; cDepartment of Molecular and Cellular Medicine, Texas A&M Health Science Center, Texas A&M University, Bryan, TX 77807; dMaterials Science and Engineering, College of Engineering, Texas A&M University, College Station, TX 77843; and eCenter for Remote Health Technologies and Systems, Texas A&M University, College Station, TX 77843 Edited by Catherine J. Murphy, University of Illinois at Urbana–Champaign, Urbana, IL, and approved April 10, 2020 (received for review August 18, 2019) Two-dimensional (2D) molybdenum disulfide (MoS2) nanomateri- Subsequently, we utilize RNA-seq to characterize the in-depth and als are an emerging class of biomaterials that are photoresponsive unbiased effect of MoS2 on the gene expression program of hMSCs. at near-infrared wavelengths (NIR). Here, we demonstrate the RNA-seq reveals the steady-state expression of the transcriptome ability of 2D MoS to modulate cellular functions of human stem 2 after the MoS2 treatment, as well as the photothermal effect of cells through photothermal mechanisms. The interaction of MoS2 MoS2 at near-infrared wavelengths (NIR). This information assists and NIR stimulation of MoS2 with human stem cells is investigated in identifying the role of MoS and NIR exposure in influencing key using whole-transcriptome sequencing (RNA-seq). -

Immune Cell Guide
Immune cell guide Antibodies Human and mouse antigens Full spectrum cell analysis Each cell holds a mystery waiting to be solved. At Thermo Fisher Scientific, we are focussed on accelerating your science and advancing meaningful discoveries by providing a comprehensive suite of solutions for the analysis of cells and their functions. Our innovative products include the Invitrogen™ Attune™ NxT Flow Cytometer, eBioscience™ flow cytometry antibodies and Super Bright conjugates, PrimeFlow™ gene expression assays, and functional assays. When you are on your quest for significant breakthroughs, we know that you never settle for average, and neither will we. Abbreviations Contents Act Activated RBC Red blood cells ASV Alternative splice variants RTK Receptor tyrosine kinase ADCC Antibody-dependent cellular cytotoxicity Tfh Folicular T helper cells B cell markers 4 AML Acute myeloid leukemia Th T helper cells APC Antigen presenting cells TF Transcription factor CD4 T cell markers 6 BM Bone marrow TM Transmembrane CCRSF Complement component receptor superfamily TM12SF 12-transmembrane spanning protein superfamily CD Cluster of differentiation TM4SF 4-transmembrane spanning protein superfamily CD8 T cell markers 8 CHO Carbohydrate moiety TM7SF 7-transmembrane spanning protein superfamily CNS Central nervous system TNFRSF TNF receptor superfamily Dendritic cell markers 10 CRSF Cytokine receptor superfamily TNFSF TNF superfamily CTL Cytotoxic T lymphocytes DC Dendritic cells Granulocyte markers 12 ECM Extracellular matrix Format abbreviations Endoth