The 5S Rdna of the Bivalve Cerastoderma Edule: Nucleotide
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

National Monitoring Program for Biodiversity and Non-Indigenous Species in Egypt
UNITED NATIONS ENVIRONMENT PROGRAM MEDITERRANEAN ACTION PLAN REGIONAL ACTIVITY CENTRE FOR SPECIALLY PROTECTED AREAS National monitoring program for biodiversity and non-indigenous species in Egypt PROF. MOUSTAFA M. FOUDA April 2017 1 Study required and financed by: Regional Activity Centre for Specially Protected Areas Boulevard du Leader Yasser Arafat BP 337 1080 Tunis Cedex – Tunisie Responsible of the study: Mehdi Aissi, EcApMEDII Programme officer In charge of the study: Prof. Moustafa M. Fouda Mr. Mohamed Said Abdelwarith Mr. Mahmoud Fawzy Kamel Ministry of Environment, Egyptian Environmental Affairs Agency (EEAA) With the participation of: Name, qualification and original institution of all the participants in the study (field mission or participation of national institutions) 2 TABLE OF CONTENTS page Acknowledgements 4 Preamble 5 Chapter 1: Introduction 9 Chapter 2: Institutional and regulatory aspects 40 Chapter 3: Scientific Aspects 49 Chapter 4: Development of monitoring program 59 Chapter 5: Existing Monitoring Program in Egypt 91 1. Monitoring program for habitat mapping 103 2. Marine MAMMALS monitoring program 109 3. Marine Turtles Monitoring Program 115 4. Monitoring Program for Seabirds 118 5. Non-Indigenous Species Monitoring Program 123 Chapter 6: Implementation / Operational Plan 131 Selected References 133 Annexes 143 3 AKNOWLEGEMENTS We would like to thank RAC/ SPA and EU for providing financial and technical assistances to prepare this monitoring programme. The preparation of this programme was the result of several contacts and interviews with many stakeholders from Government, research institutions, NGOs and fishermen. The author would like to express thanks to all for their support. In addition; we would like to acknowledge all participants who attended the workshop and represented the following institutions: 1. -

Age Structure, Growth and Shell Form of Cerastoderma Glaucum (Bivalvia
Age structure, growth and shell form of Cerastoderma glaucum (Bivalvia: Cardiidae) from El Mellah lagoon, Algeria Lamia Bensaâd-Bendjedid, Saber Belhaoues, Asma Kerdoussi, Naouel Djebbari, Mardja Tahri, Mourad Bensouilah Laboratory of Ecobiology for Marine Environment and Coastlines, Faculty of Science, Badji Mokhtar University, Annaba 23000, Algeria. Corresponding author: L. Bensaâd-Benjedid, [email protected] Abstract. Cerastoderma glaucum (Poiret, 1789) the lagoon specialist cockle, represents one of the most abundant molluscs in El Mellah coastal lagoon (northeastern Algeria); several morphological and biological characteristics of this species were investigated over a period of 18 months (January 2014 to June 2015). Analysis of allometry and different morphometric indices indicate that this population tends to assume a globular shell shape which becomes elongated as the cockle grows. From length-frequency data examination using software FiSAT II and VONBIT for Excel, age was ranged from the first to the third year old, asymptotic length (L∞) was estimated to 48.061 mm and growth coefficient (K) was 0.71 -1 yr . The growth performance index (Φ′) and theoretical life span (tmax) were 3.21 and 4.23 yr respectively, total mortality rate (Z) obtained was 1.94 yr-1. Monthly variations in condition index were noticed, two sharp falls due spawning events were observed, the first spawning occurred between March and June and, the second one took place from August to October. The findings of the current study can be utilized to guide future research in various fields as pathology, biomonitoring and environmental management of this species. Key Words: Cerastoderma glaucum, shell shape, growth, Condition index, Algeria. -

Download PDF Version
MarLIN Marine Information Network Information on the species and habitats around the coasts and sea of the British Isles Lagoon cockle (Cerastoderma glaucum) MarLIN – Marine Life Information Network Biology and Sensitivity Key Information Review Nicola White 2002-07-15 A report from: The Marine Life Information Network, Marine Biological Association of the United Kingdom. Please note. This MarESA report is a dated version of the online review. Please refer to the website for the most up-to-date version [https://www.marlin.ac.uk/species/detail/1315]. All terms and the MarESA methodology are outlined on the website (https://www.marlin.ac.uk) This review can be cited as: White, N. 2002. Cerastoderma glaucum Lagoon cockle. In Tyler-Walters H. and Hiscock K. (eds) Marine Life Information Network: Biology and Sensitivity Key Information Reviews, [on-line]. Plymouth: Marine Biological Association of the United Kingdom. DOI https://dx.doi.org/10.17031/marlinsp.1315.1 The information (TEXT ONLY) provided by the Marine Life Information Network (MarLIN) is licensed under a Creative Commons Attribution-Non-Commercial-Share Alike 2.0 UK: England & Wales License. Note that images and other media featured on this page are each governed by their own terms and conditions and they may or may not be available for reuse. Permissions beyond the scope of this license are available here. Based on a work at www.marlin.ac.uk (page left blank) Date: 2002-07-15 Lagoon cockle (Cerastoderma glaucum) - Marine Life Information Network See online review for distribution map Three Cerastoderma glaucum with siphons extended. Distribution data supplied by the Ocean Photographer: Dennis R. -

Certification Report for Burry Inlet
Moody Marine Ltd Burry Inlet Cockle Fishery: Public Certification Report MOODY MARINE LTD Ref: 82001 v5 Author(s): T Holt, A Hough Certification Report for Burry Inlet Cockle Fishery Client: South Wales Sea Fishery Committee Certification Body: Client Contact: Moody Marine Ltd Director: Mr P Coates Moody International Certification South Wales Sea Fisheries Committee Salisbury House Queens Buildings Stephenson’s Way Cambrian Place Wyvern Business Park Swansea. SA1 1TW Derby. DE21 6LY UK UK Tel: +44 (0) 1704 834644 01792 654466 Fax: +44 (0) 1332 675152 01792 645987 FN 07/019 82001 v4 1 Moody Marine Ltd Burry Inlet Cockle Fishery: Public Certification Report CONTENTS 1. INTRODUCTION..................................................................................................................................... 3 1.1 THE FISHERY PROPOSED FOR CERTIFICATION...................................................................................... 3 1.2 REPORT STRUCTURE AND ASSESSMENT PROCESS .............................................................................. 3 1.3 INFORMATION SOURCES USED ............................................................................................................ 4 2 BACKGROUND TO THE FISHERY .................................................................................................... 6 2.1 BIOLOGY OF THE TARGET SPECIES ..................................................................................................... 6 2.2 HISTORY OF THE FISHERY ................................................................................................................. -

Spatial Variability in Recruitment of an Infaunal Bivalve
Spatial Variability in Recruitment of an Infaunal Bivalve: Experimental Effects of Predator Exclusion on the Softshell Clam (Mya arenaria L.) along Three Tidal Estuaries in Southern Maine, USA Author(s): Brian F. Beal, Chad R. Coffin, Sara F. Randall, Clint A. Goodenow Jr., Kyle E. Pepperman, Bennett W. Ellis, Cody B. Jourdet and George C. Protopopescu Source: Journal of Shellfish Research, 37(1):1-27. Published By: National Shellfisheries Association https://doi.org/10.2983/035.037.0101 URL: http://www.bioone.org/doi/full/10.2983/035.037.0101 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Journal of Shellfish Research, Vol. 37, No. 1, 1–27, 2018. SPATIAL VARIABILITY IN RECRUITMENT OF AN INFAUNAL BIVALVE: EXPERIMENTAL EFFECTS OF PREDATOR EXCLUSION ON THE SOFTSHELL CLAM (MYA ARENARIA L.) ALONG THREE TIDAL ESTUARIES IN SOUTHERN MAINE, USA 1,2 3 2 3 BRIAN F. -

HETA ROUSI: Zoobenthos As Indicators of Marine Habitats in the Northern Baltic
Heta Rousi Zoobenthos as indicators of marine Heta Rousi | habitats in the northern Baltic Sea of marine as indicators habitats in the northernZoobenthos Baltic Sea Heta Rousi This thesis describes how physical and chemical environmental variables impact zoobenthic species distribution in the northern Baltic Sea and how dis- Zoobenthos as indicators of marine tinct zoobenthic species indicate different marine benthic habitats. The thesis inspects the effects of habitats in the northern Baltic Sea depth, sediment type, temperature, salinity, oxy- gen, nutrients as well as topographical and geo- logical factors on zoobenthos on small and large temporal and spatial scales. | 2020 ISBN 978-952-12-3944-1 Heta Rousi Född 1979 Studier och examina Magister vid Helsingfors Universitet 2006 Licentiat vid Åbo Akademi 2013 Doktorsexamen vid Åbo Akademi 2020 Institutionen för miljö- och marinbiologi, Åbo Akademi ZOOBENTHOS AS INDICATORS OF MARINE HABITATS IN THE NORTHERN BALTIC SEA HETA ROUSI Environmental and Marine Biology Faculty of Science and Engineering Åbo Akademi University Finland, 2020 SUPERVISED BY PRE-EXAMINED BY Professor Erik Bonsdorff Research Professor (Supervisor & Examiner) Markku Viitasalo Åbo Akademi University Finnish Environment Institute Faculty of Science and Engineering Sustainable Use of the Marine Areas Environmental and Marine Biology Latokartanonkaari 11 Artillerigatan 6 00790 Helsinki 20520 Åbo Finland Finland Professor Emeritus Ilppo Vuorinen CO-SUPERVISOR University of Turku Adjunct Professor Faculty of Science and Engineering Samuli Korpinen Itäinen Pitkäkatu 4 Finnish Environment Institute 20520 Turku Marine Management Finland Latokartanonkaari 11 00790 Helsinki FACULTY OPPONENT Finland Associate Professor Urszula Janas SUPERVISING AT THE University of Gdansk LICENCIATE PHASE Institute of Oceanography Assistant Professor Al. -

Digenean Fauna of Cerastoderma Glaucum (Veneroidae, Cardidae) from Tunisian Coasts
Bull. Eur. Ass. Fish Pathol., 31(1) 2011, 4 Digenean fauna of Cerastoderma glaucum (Veneroidae, Cardidae) from Tunisian coasts L. Gargouri Ben Abdallah1*, R. Antar1, K. Hosni2, N. Trigui El Menif2 and F. Maamouri1 1 Reasearch Unit Animal Bio-Ecology and Systematic Evolutionary, Faculty of Sciences, Tunis, University of Tunis El Manar, 2092, Tunis, Tunisia; 2 Laboratory of Environment Bio-monitoring (LBE), Faculty of Sciences, Bizerte, University 7th November Carthage, 7021 Zarzouna, Bizerte,Tunisia Abstract The study conducted on the cockle Cerastoderma glaucum from Tunisian coasts (Bizerte lagoon and Gulf of Gabes) showed the presence of two digenean species (sporocyst and cercaria of Bucephalus minimus and metacercaria of Meiogymnophallus strigatus). A description of these larvae and the behaviour of the naturally emerging cercaria are reported. The examination of the frequency of B. minimus and M. strigatus reveals some seasonal variation. This variation becomes more significant by comparing the sites. B. minimus has the highest frequency in the Bizerte lagoon, by contrast to that of M. strigatus which is the highest in the Gulf of Gabes. This variation is statistically significant. These results indicate that B. minimus and M. strigatus can be considered as biological indicators making it possible to predict the capture area of the cockle. Introduction The cockle, Cerastoderma glaucum, which has process of regulating of the natural popula- a significant economic value, is commonly tions of marine organisms, is a theme which exploited in several European countries. In was the subject of little attention because of the Tunisia, in spite of its abundance in several durability of the relation parasite-host (Combes, sites, in lagoons as well as in the sea, this species 1995). -

Tayside, Central and Fife Tayside, Central and Fife
Detail of the Lower Devonian jawless, armoured fish Cephalaspis from Balruddery Den. © Perth Museum & Art Gallery, Perth & Kinross Council Review of Fossil Collections in Scotland Tayside, Central and Fife Tayside, Central and Fife Stirling Smith Art Gallery and Museum Perth Museum and Art Gallery (Culture Perth and Kinross) The McManus: Dundee’s Art Gallery and Museum (Leisure and Culture Dundee) Broughty Castle (Leisure and Culture Dundee) D’Arcy Thompson Zoology Museum and University Herbarium (University of Dundee Museum Collections) Montrose Museum (Angus Alive) Museums of the University of St Andrews Fife Collections Centre (Fife Cultural Trust) St Andrews Museum (Fife Cultural Trust) Kirkcaldy Galleries (Fife Cultural Trust) Falkirk Collections Centre (Falkirk Community Trust) 1 Stirling Smith Art Gallery and Museum Collection type: Independent Accreditation: 2016 Dumbarton Road, Stirling, FK8 2KR Contact: [email protected] Location of collections The Smith Art Gallery and Museum, formerly known as the Smith Institute, was established at the bequest of artist Thomas Stuart Smith (1815-1869) on land supplied by the Burgh of Stirling. The Institute opened in 1874. Fossils are housed onsite in one of several storerooms. Size of collections 700 fossils. Onsite records The CMS has recently been updated to Adlib (Axiel Collection); all fossils have a basic entry with additional details on MDA cards. Collection highlights 1. Fossils linked to Robert Kidston (1852-1924). 2. Silurian graptolite fossils linked to Professor Henry Alleyne Nicholson (1844-1899). 3. Dura Den fossils linked to Reverend John Anderson (1796-1864). Published information Traquair, R.H. (1900). XXXII.—Report on Fossil Fishes collected by the Geological Survey of Scotland in the Silurian Rocks of the South of Scotland. -

Behind Anemone Lines: Determining the Environmental Drivers Influencing Lagoonal Benthic Communities, with Special Reference to the Anemone Nematostella Vectensis
Behind Anemone Lines: Determining the environmental drivers influencing lagoonal benthic communities, with special reference to the anemone Nematostella vectensis. by Jessica R. Bone Bournemouth University December 2018 Copyright Statement This copy of the thesis has been supplied on condition that anyone who consults it is understood to recognize that its copyright rests with its author and due acknowledgement must always be made of the use of any material contained in, or derived from, this thesis. i Behind Anemone Lines: Determining the environmental drivers influencing lagoonal benthic communities, with special reference to the anemone Nematostella vectensis. Jess R. Bone Abstract Climate change induced sea level rise and increase in associated storms is impacting the coastal zone worldwide. Lagoons are a transitional ecosystem on the coast that are threatened with habitat loss due to ingress of seawater, though conversely this also represents an opportunity for lagoon habitat creation. It is important to quantify the spatio-temporal trends of macrozoobenthic communities and abiotic factors to determine the ecological health of lagoon sites. Such information will ensure optimal and adaptive management of these rare and protected ecosystems. This thesis examines the spatial distribution of macrozoobenthic assemblages and the abiotic and biotic factors that may determine their abundance, richness and distribution at tidally restricted urban lagoon at Poole Park on the south coast of England. The macrozoobenthic assemblages were sampled using a suction corer during a spatially comprehensive survey in November 2017, in addition to aquatic and sediment variables such as salinity, temperature, organic matter content and silt content. Species richness and density were significantly lower in areas of high organic matter and silt content, indicative of hostile conditions. -
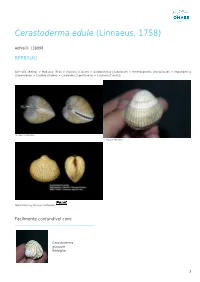
Cerastoderma Edule (Linnaeus, 1758)
Cerastoderma edule (Linnaeus, 1758) AphiaID: 138998 BERBIGÃO Animalia (Reino) > Mollusca (Filo) > Bivalvia (Classe) > Autobranchia (Subclasse) > Heteroconchia (Infraclasse) > Imparidentia (Superordem) > Cardiida (Ordem) > Cardioidea (Superfamilia) > Cardiidae (Familia) © Vasco Ferreira © Vasco Ferreira Natural History Museum Rotterdam Facilmente confundível com: Cerastoderma glaucum Berbigão 1 Estatuto de Conservação Sinónimos Cardium belgicum De Malzine, 1867 Cardium crenulatum Lamarck, 1819 Cardium edule Linnaeus, 1758 Cardium edule burchanae Girscher, 1938 Cardium edule var. batesoni Bucquoy, Dautzenberg & Dollfus, 1892 Cardium edule var. loppensi Mars, 1951 Cardium edule var. maculata Dautzenberg, 1890 Cardium edule var. major Bucquoy, Dautzenberg & Dollfus, 1892 Cardium edule var. mareotica Pallary, 1912 Cardium edule var. regularis Pallary, 1900 Cardium edule var. sibenicensis Brusina, 1870 Cardium mercatorium Coen, 1915 Cardium nunninkae Lucas, 1984 Cardium obtritum Locard, 1886 Cardium quadrarium Reeve, 1845 Cardium vulgare da Costa, 1778 Cardium vulgatum Tryon, 1872 Cerastoderma edule var. sinicola Lacourt, 1974 Cerastoderma nunninkae Lucas, 1984 Referências basis of record Gofas, S.; Le Renard, J.; Bouchet, P. (2001). Mollusca. in: Costello, M.J. et al. (eds), European Register of Marine Species: a check-list of the marine species in Europe and a bibliography of guides to their identification. Patrimoines Naturels. 50: 180-213. [details] additional source Poorten, J.J. ter, 2005. Outline of a systematic index – Recent Cardiidae (Lamarck, 1809). VISAYA net. (Updated 2009 for WoRMS), available online at http://www.conchology.be/en/shelltopics/visaya-net/date.php?year=2005 [details] 2 ecology source Coscia, I., P.E. Robins, J.S. Porter, S.K. Malham & J.E. Ironside. (2013). Modelled larval dispersal and measured gene flow: seascape genetics of the common cockle Cerastoderma edule in the southern Irish Sea. -

Ruditapes Philippinarum (Adams and Reeve, 1850) and Trophic Niche Overlap with Native Species
Aquatic Invasions (2019) Volume 14, Issue 4: 638–655 CORRECTED PROOF Research Article Food sources of the non-indigenous bivalve Ruditapes philippinarum (Adams and Reeve, 1850) and trophic niche overlap with native species Ester Dias1,*, Paula Chainho2, Cristina Barrocas-Dias3,4 and Helena Adão5 1CIMAR/CIIMAR –Interdisciplinary Centre of Marine and Environmental Research, University of Porto, Terminal de Cruzeiros do Porto de Leixões, Av. General Norton de Matos, 4450-208 Matosinhos, Portugal 2MARE, Faculdade de Ciências da Universidade de Lisboa, Campo Grande, 1749-016 Lisboa, Portugal 3HERCULES Laboratory, University of Évora, Largo Marquês de Marialva, 8, 7000-809 Évora, Portugal 4Chemistry Department, School of Sciences and Technology, Rua Romão Ramalho 59, Évora, Portugal 5MARE- University of Évora, School of Sciences and Technology, Apartado 94, 7005-554 Évora, Portugal Author e-mails: [email protected] (ED), [email protected] (PC), [email protected] (CBD), [email protected] (HA) *Corresponding author Citation: Dias E, Chainho P, Barrocas- Dias C, Adão H (2019) Food sources of the Abstract non-indigenous bivalve Ruditapes philippinarum (Adams and Reeve, 1850) The Manila clam, Ruditapes philippinarum (Adams and Reeve, 1850), was introduced and trophic niche overlap with native in many estuaries along the Atlantic and Mediterranean coasts for fisheries and species. Aquatic Invasions 14(4): 638–655, aquaculture, being one of the top five most commercially valuable bivalve species https://doi.org/10.3391/ai.2019.14.4.05 worldwide. In Portugal, the colonization of the Tagus estuary by this species Received: 8 February 2019 coincided with a significant decrease in abundance of the native R. -

Macrobenthic Assemblage Structure and Distribution at the Boojagh Marine National Park, Southern Caspian Sea, Iran
Iranian Journal of Fisheries Sciences 19(2) 748-767 2020 DOI: 10.22092/ijfs.2020.120829 Macrobenthic assemblage structure and distribution at the Boojagh Marine National Park, Southern Caspian Sea, Iran. Bahrebar S.¹; Negarestan H.²*; Maghsoudlo A.³; Danehkar A.4 Received: December 2016 Accepted: Febtuary 2017 Abstract Although macrobenthic assemblages are considered as major players in many ecosystems around the world, the ecology of Caspian Sea macrobenthos is currently understudied. This study describes the species composition and quantitative distribution of macrobenthos in the southern Caspian Sea and relates the distribution to seasonal changes at three depths (1, 5 and 10 meters) on the Boojagh Marine National Park (BMNP) coast in the southern Caspian Sea between the summers of 2015 and 2016. To investigate the distribution of macrobenthos in BMNP, the data of 450 samples were analyzed. In this study sixteen species were identified: Cerastoderma glaucum, Mytilaster lineatus, Pyrgula grimmi, Anisus kolesnikovi, Stenogammarus carausui, Paraniphargoides motasi, Onisimus caspius, Pterocuma pectinatum, Pterocuma sowinskyi, Pseudocuma (Stenocuma) gracile, Nais sp., Hypania invalida, Manayunkia caspica, Streblospio gynobranchiata, Hediste diversicolor, Amphibalanus improvisus. Among them, the non-indigenous C. glaucum was the dominant species, accounting for 27% of the total abundance and in descending order P. grimmi with 14.4%, A. improvisus with 8.7%, M. lineatus with 7.9%, Nais sp. with 7.5%, N. carausui with 5.2%, P. motasi with 5%, S. gynobranchiata with 4.5%, H. invalida with 5%, M. Caspica with 3.1%, P. sowinskyi with 2.5%, O. caspius with 2.4%, A. kolesnikovi and H. diversicolor with 1.8%, S.