Title Microbiota in the Coelomic Fluid of Two Common Coastal Starfish
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Structural Characterization of Helicobacter Pylori Proteins Contributing to Stomach Colonization
Università degli Studi di Padova Dipartimento di Biologia Scuola di Dottorato di Ricerca in Bioscienze e Biotecnologie Indirizzo: Biotecnologie Ciclo XXVIII STRUCTURAL CHARACTERIZATION OF HELICOBACTER PYLORI PROTEINS CONTRIBUTING TO STOMACH COLONIZATION Direttore della Scuola: Ch.mo Prof. Paolo Bernardi Coordinatore di Indirizzo: Ch.ma Prof.ssa Fiorella Lo Schiavo Supervisore: Ch.mo Prof. Giuseppe Zanotti Dottorando: Maria Elena Compostella 31 Gennaio 2016 Università degli Studi di Padova Department of Biology School of Biosciences and Biotechnology Curriculum: Biotechnology XXVIII Cycle STRUCTURAL CHARACTERIZATION OF HELICOBACTER PYLORI PROTEINS CONTRIBUTING TO STOMACH COLONIZATION Director of the Ph.D. School: Ch.mo Prof. Paolo Bernardi Coordinator of the Curriculum: Ch.ma Prof.ssa Fiorella Lo Schiavo Supervisor: Ch.mo Prof. Giuseppe Zanotti Ph.D. Candidate: Maria Elena Compostella 31 January 2016 Contents ABBREVIATIONS AND SYMBOLS IV SUMMARY 9 SOMMARIO 15 1. INTRODUCTION 21 1.1 HELICOBACTER PYLORI 23 1.2 GENETIC VARIABILITY 26 1.2.1 GENOME COMPARISON 26 1.2.1.1 HELICOBACTER PYLORI 26695 26 1.2.1.2 HELICOBACTER PYLORI J99 28 1.2.2 CORE GENOME 30 1.2.3 MECHANISMS GENERATING GENETIC VARIABILITY 31 1.2.3.1 MUTAGENESIS 32 1.2.3.2 RECOMBINATION 35 1.2.4 HELICOBACTER PYLORI AS A “QUASI SPECIES” 37 1.2.5 CLASSIFICATION OF HELICOBACTER PYLORI STRAINS 38 1.3 EPIDEMIOLOGY 40 1.3.1 INCIDENCE AND PREVALENCE OF HELICOBACTER PYLORI INFECTION 40 1.3.2 SOURCE AND TRANSMISSION 42 1.4 ADAPTATION AND GASTRIC COLONIZATION 47 1.4.1 ACID ADAPTATION 49 1.4.2 MOTILITY AND CHEMIOTAXIS 60 1.4.3 ADHESION 65 1.5 PATHOGENESIS AND VIRULENCE FACTORS 72 1.5.1 VACUOLATING CYTOTOXIN A 78 1.5.2 CAG PATHOGENICITY ISLAND AND CYTOTOXIN-ASSOCIATED GENE A 83 1.5.3 NEUTROPHIL-ACTIVATING PROTEIN 90 1.6 HELICOBACTER PYLORI AND GASTRODUODENAL DISEASES 92 1.7 ERADICATION AND POTENTIAL BENEFITS 97 2. -

Food Or Beverage Product, Or Probiotic Composition, Comprising Lactobacillus Johnsonii 456
(19) TZZ¥¥¥ _T (11) EP 3 536 328 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 11.09.2019 Bulletin 2019/37 A61K 35/74 (2015.01) A61K 35/66 (2015.01) A61P 35/00 (2006.01) (21) Application number: 19165418.5 (22) Date of filing: 19.02.2014 (84) Designated Contracting States: • SCHIESTL, Robert, H. AL AT BE BG CH CY CZ DE DK EE ES FI FR GB Encino, CA California 91436 (US) GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO • RELIENE, Ramune PL PT RO RS SE SI SK SM TR Los Angeles, CA California 90024 (US) • BORNEMAN, James (30) Priority: 22.02.2013 US 201361956186 P Riverside, CA California 92506 (US) 26.11.2013 US 201361909242 P • PRESLEY, Laura, L. Santa Maria, CA California 93458 (US) (62) Document number(s) of the earlier application(s) in • BRAUN, Jonathan accordance with Art. 76 EPC: Tarzana, CA California 91356 (US) 14753847.4 / 2 958 575 (74) Representative: Müller-Boré & Partner (71) Applicant: The Regents of the University of Patentanwälte PartG mbB California Friedenheimer Brücke 21 Oakland, CA 94607 (US) 80639 München (DE) (72) Inventors: Remarks: • YAMAMOTO, Mitsuko, L. This application was filed on 27-03-2019 as a Alameda, CA California 94502 (US) divisional application to the application mentioned under INID code 62. (54) FOOD OR BEVERAGE PRODUCT, OR PROBIOTIC COMPOSITION, COMPRISING LACTOBACILLUS JOHNSONII 456 (57) The present invention relates to food products, beverage products and probiotic compositions comprising Lacto- bacillus johnsonii 456. EP 3 536 328 A1 Printed by Jouve, 75001 PARIS (FR) EP 3 536 328 A1 Description CROSS-REFERENCE TO RELATED APPLICATIONS 5 [0001] This application claims the benefit of U.S. -

In Silico Evolutionary Analysis of Helicobacter Pylori Outer Membrane Phospholipase a (OMPLA) Hilde S Vollan1*, Tone Tannæs1, Yoshio Yamaoka2 and Geir Bukholm3,4
Vollan et al. BMC Microbiology 2012, 12:206 http://www.biomedcentral.com/1471-2180/12/206 RESEARCH ARTICLE Open Access In silico evolutionary analysis of Helicobacter pylori outer membrane phospholipase A (OMPLA) Hilde S Vollan1*, Tone Tannæs1, Yoshio Yamaoka2 and Geir Bukholm3,4 Abstract Background: In the past decade, researchers have proposed that the pldA gene for outer membrane phospholipase A (OMPLA) is important for bacterial colonization of the human gastric ventricle. Several conserved Helicobacter pylori genes have distinct genotypes in different parts of the world, biogeographic patterns that can be analyzed through phylogenetic trees. The current study will shed light on the importance of the pldA gene in H. pylori. In silico sequence analysis will be used to investigate whether the bacteria are in the process of preserving, optimizing, or rejecting the pldA gene. The pldA gene will be phylogenetically compared to other housekeeping (HK) genes, and a possible origin via horizontal gene transfer (HGT) will be evaluated through both intra- and inter- species evolutionary analyses. Results: In this study, pldA gene sequences were phylogenetically analyzed and compared with a large reference set of concatenated HK gene sequences. A total of 246 pldA nucleotide sequences were used; 207 were from Norwegian isolates, 20 were from Korean isolates, and 19 were from the NCBI database. Best-fit evolutionary models were determined with MEGA5 ModelTest for the pldA (K80 + I + G) and HK (GTR + I + G) sequences, and maximum likelihood trees were constructed. Both HK and pldA genes showed biogeographic clustering. Horizontal gene transfer was inferred based on significantly different GC contents, the codon adaptation index, and a phylogenetic conflict between a tree of OMPLA protein sequences representing 171 species and a tree of the AtpA HK protein for 169 species. -

Genomics of Helicobacter Species 91
Genomics of Helicobacter Species 91 6 Genomics of Helicobacter Species Zhongming Ge and David B. Schauer Summary Helicobacter pylori was the first bacterial species to have the genome of two independent strains completely sequenced. Infection with this pathogen, which may be the most frequent bacterial infec- tion of humanity, causes peptic ulcer disease and gastric cancer. Other Helicobacter species are emerging as causes of infection, inflammation, and cancer in the intestine, liver, and biliary tract, although the true prevalence of these enterohepatic Helicobacter species in humans is not yet known. The murine pathogen Helicobacter hepaticus was the first enterohepatic Helicobacter species to have its genome completely sequenced. Here, we consider functional genomics of the genus Helico- bacter, the comparative genomics of the genus Helicobacter, and the related genera Campylobacter and Wolinella. Key Words: Cytotoxin-associated gene; H-Proteobacteria; gastric cancer; genomic evolution; genomic island; hepatobiliary; peptic ulcer disease; type IV secretion system. 1. Introduction The genus Helicobacter belongs to the family Helicobacteriaceae, order Campylo- bacterales, and class H-Proteobacteria, which is also known as the H subdivision of the phylum Proteobacteria. The H-Proteobacteria comprise of a relatively small and recently recognized line of descent within this extremely large and phenotypically diverse phy- lum. Other genera that colonize and/or infect humans and animals include Campylobac- ter, Arcobacter, and Wolinella. These organisms are all microaerophilic, chemoorgano- trophic, nonsaccharolytic, spiral shaped or curved, and motile with a corkscrew-like motion by means of polar flagella. Increasingly, free living H-Proteobacteria are being recognized in a wide range of environmental niches, including seawater, marine sedi- ments, deep-sea hydrothermal vents, and even as symbionts of shrimp and tubeworms in these environments. -

Co-Infection Associated with Diarrhea in a Colony of <I>Scid
Laboratory Animal Science Vol 48, No 5 Copyright 1998 October 1998 by the American Association for Laboratory Animal Science Helicobacter bilis/Helicobacter rodentium Co-Infection Associated with Diarrhea in a Colony of scid Mice Nirah H. Shomer,* Charles A. Dangler, Robert P. Marini, and James G. Fox† Abstract _ An outbreak of diarrhea spanning 3 months occurred in a breeding colony of scid/Trp53 knockout mice. Approximately a third of the 150 mice were clinically affected, with signs ranging from mucoid or watery diarrhea to severe hemorrhagic diarrhea with mortality. Helicobacter bilis and the newly recognized urease-negative organ- ism H. rodentium were isolated from microaerobic culture of feces or cecal specimens from affected mice. Dual infection with H. bilis and H. rodentium were confirmed by culture and polymerase chain reaction (PCR) in several animals. Both Helicobacter species rapidly colonized immunocompetent sentinel mice exposed to bedding from cages containing affected mice, but the sentinel remained asymptomatic. Mice with diarrhea had multifocal to segmental proliferative typhlitis, colitis, and proctitis. Several affected mice had multifocal mucosal necrosis with a few focal ulcers in the cecum, colon, and rectum. Mice with diarrhea were treated with antibiotic food wafers (1.5 mg of amoxicillin, 0.69 mg of metronidazole, and 0.185 mg of bismuth/mouse per day) previously shown to eradi- cate H. hepaticus in immunocompetent mice. Antibiotic treatment resulted in resolution of diarrhea, but not eradication of H. bilis and H. rodentium; mice continued to have positive PCR results after a 2-week treatment regimen, and clinical signs of diarrhea returned in some mice when treatment was suspended. -

Evolutionary Dynamics of the Vapd Gene in Helicobacter Pylori and Its
ISSN Online: 2372-0956 Symbiosis www.symbiosisonlinepublishing.com Research Article SOJ Microbiology & Infectious Diseases Open Access Evolutionary dynamics of the vapD gene in Helicobacter pylori and its wide distribution among bacterial phyla Gabriela Delgado-Sapién1, Rene Cerritos-Flores2, Alejandro Flores-Alanis1, José L Méndez1, Alejandro Cravioto1, Rosario Morales-Espinosa1* *1Laboratorio de Genómica Bacteriana, Departamento de Microbiología y Parasitología, Facultad de Medicina, Universidad Nacional Autónoma de México, Mexico City, México 04510. 2Centro de Investigación en Políticas, Población y Salud, Facultad de Medicina, Universidad Nacional Autónoma de México, Mexico City, México 04510. Received: 12th August , 2020; Accepted: 15th November 2020 ; Published: 03rd December, 2020 *Corresponding author: RosarioMorales-Espinosa, PhD, MD, Laboratorio de Genómica Bacteriana, Departamento de Microbiología y Parasi- tología. Universidad Nacional Autónoma de México. Avenida Universidad 3000, Colonia Ciudad Universitaria, Delegación Coyoacán, C.P. 04510, México City, México.Tel.: +525 5523 2135; Fax: +525 5623 2114 E-mail: [email protected] factors [1,2,3]. Genetic diversity is seen among H. pylori strains Abstract from different origins and ethnic populations, as well as within The vapD gene is present in microorganisms from different phyla H. pylori populations within a single stomach. It is well known and encodes for the virulence-associated protein D (VapD). In some that H. pylori is a highly recombinant microorganism [4-8] and microorganisms, it has been suggested that vapD participates in either a natural transformant, which explains its genomic variability protecting the bacteria from respiratory burst within the macrophage and diversity that favour a better adaptive capacity and its or in facilitating the persistence of the microorganism within the permanence on the gastric mucosa for decades. -

List of the Pathogens Intended to Be Controlled Under Section 18 B.E
(Unofficial Translation) NOTIFICATION OF THE MINISTRY OF PUBLIC HEALTH RE: LIST OF THE PATHOGENS INTENDED TO BE CONTROLLED UNDER SECTION 18 B.E. 2561 (2018) By virtue of the provision pursuant to Section 5 paragraph one, Section 6 (1) and Section 18 of Pathogens and Animal Toxins Act, B.E. 2558 (2015), the Minister of Public Health, with the advice of the Pathogens and Animal Toxins Committee, has therefore issued this notification as follows: Clause 1 This notification is called “Notification of the Ministry of Public Health Re: list of the pathogens intended to be controlled under Section 18, B.E. 2561 (2018).” Clause 2 This Notification shall come into force as from the following date of its publication in the Government Gazette. Clause 3 The Notification of Ministry of Public Health Re: list of the pathogens intended to be controlled under Section 18, B.E. 2560 (2017) shall be cancelled. Clause 4 Define the pathogens codes and such codes shall have the following sequences: (1) English alphabets that used for indicating the type of pathogens are as follows: B stands for Bacteria F stands for fungus V stands for Virus P stands for Parasites T stands for Biological substances that are not Prion R stands for Prion (2) Pathogen risk group (3) Number indicating the sequence of each type of pathogens Clause 5 Pathogens intended to be controlled under Section 18, shall proceed as follows: (1) In the case of being the pathogens that are utilized and subjected to other law, such law shall be complied. (2) Apart from (1), the law on pathogens and animal toxin shall be complied. -

Genomic Analysis of Helicobacter Himalayensis Sp. Nov. Isolated from Marmota Himalayana
Genomic analysis of Helicobacter himalayensis sp. nov. isolated from Marmota himalayana Shouhui Hu Peking University Shougang Hospital Lina Niu Hainan Medical University Lei Wu Peking University Shougang Hospital Xiaoxue Zhu Peking University Shougang Hospital Yu Cai Peking University Shougang Hospital Dong Jin Chinese Center for Disease Control and Prevention Linlin Yan Peking University Shougang Hospital Fan Zhao ( [email protected] ) Peking University Shougang Hospital https://orcid.org/0000-0002-8164-5016 Research article Keywords: Helicobacter, Comparative genomics, Helicobacter himalayensis, Virulence factor Posted Date: December 1st, 2020 DOI: https://doi.org/10.21203/rs.3.rs-55448/v3 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published on November 23rd, 2020. See the published version at https://doi.org/10.1186/s12864-020-07245-y. Page 1/18 Abstract Background: Helicobacter himalayensis was isolated from Marmota himalayana in the Qinghai-Tibet Plateau, China, and is a new non-H. pylori species, with unclear taxonomy, phylogeny, and pathogenicity. Results: A comparative genomic analysis was performed between the H. himalayensis type strain 80(YS1)T and other the genomes of Helicobacter species present in the National Center for Biotechnology Information (NCBI) database to explore the molecular evolution and potential pathogenicity of H. himalayensis. H. himalayensis 80(YS1)T formed a clade with H. cinaedi and H. hepaticus that was phylogenetically distant from H. pylori. The H. himalayensis genome showed extensive collinearity with H. hepaticus and H. cinaedi. However, it also revealed a low degree of genome collinearity with H. -

Helicobacter Pylori: Comparative Genomics and Structure-Function Analysis of the Flagellum Biogenesis Protein HP0958
UCC Library and UCC researchers have made this item openly available. Please let us know how this has helped you. Thanks! Title Helicobacter pylori: comparative genomics and structure-function analysis of the flagellum biogenesis protein HP0958 Author(s) de Lacy Clancy, Ceara A. Publication date 2014 Original citation de Lacy Clancy, C. A. 2014. Helicobacter pylori: comparative genomics and structure-function analysis of the flagellum biogenesis protein HP0958. PhD Thesis, University College Cork. Type of publication Doctoral thesis Rights © 2014, Ceara A. De Lacy Clancy. http://creativecommons.org/licenses/by-nc-nd/3.0/ Item downloaded http://hdl.handle.net/10468/1684 from Downloaded on 2021-10-10T14:33:51Z Helicobacter pylori: Comparative genomics and structure-function analysis of the flagellum biogenesis protein HP0958 A Thesis Presented in Partial Fulfilment of the Requirements for the Degree of Doctor of Philosophy by Ceara de Lacy Clancy, B.Sc. School of Microbiology National University of Ireland, Cork Supervisor: Prof. Paul W. O’Toole Head of School of Microbiology: Prof. Gerald Fitzgerald February 2014 Table of Contents Table of Contents Abstract ........................................................................................................................ i Chapter 1 Literature Review....................................................................................................... 1 1 Helicobacter pylori .................................................................................................. 2 1.1 Discovery -
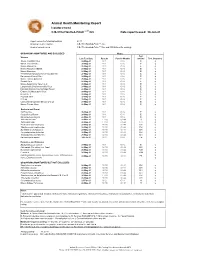
2021 ARC Health Report
Animal Health Monitoring Report Isolator reared C.B-17/IcrHanHsd-Prkdc scid /Arc Date report issued: 30-Jun-21 Report covers the following isolators: RI 27 Strains present in isolator: C.B-17/IcrHanHsd-Prkdc scid /Arc Strain of animal tested: C.B-17/IcrHanHsd-Prkdc scid /Arc and ICR Outbred for serology ORGANISMS MONITORED AND EXCLUDED Mouse Test Viruses Last Test Date Results Past 18 Months method Test frequency Mouse Hepatitis Virus 26-May-21 0 / 1 0 / 6 E q Minute Virus of Mice 26-May-21 0 / 1 0 / 6 E q Mouse Parvovirus 26-May-21 0 / 1 0 / 6 E q Murine Rotavirus (EDIM) 26-May-21 0 / 1 0 / 6 E q Mouse Norovirus 26-May-21 0 / 1 0 / 6 E q Theiler's Encephalomyelitis Virus (GD VII) 26-May-21 0 / 1 0 / 6 E q Pneumonia Virus of Mice 26-May-21 0 / 1 0 / 6 E q Murine Cytomegalovirus 26-May-21 0 / 1 0 / 6 E q Sendai Virus 26-May-21 0 / 1 0 / 6 E q Mouse Adenovirus Type 1 & 2 26-May-21 0 / 1 0 / 6 E q Lymphocytic Choriomeningitis Virus 26-May-21 0 / 1 0 / 6 E q Hantaan (Korean Haemorrhagic Fever) 26-May-21 0 / 1 0 / 6 E q Ectromelia (Mousepox) Virus 26-May-21 0 / 1 0 / 6 E q Reovirus -3 26-May-21 0 / 1 0 / 6 E q Polyoma Virus 26-May-21 0 / 1 0 / 6 E q K Virus 26-May-21 0 / 1 0 / 6 E q Lactic Dehydrogenase Elevating Virus 26-May-21 0 / 1 0 / 6 E q Mouse Thymic Virus 26-May-21 0 / 1 0 / 6 E q 00-Jan-00 Bacteria and Fungi CAR bacillus 26-May-21 0 / 1 0 / 6 E q Clostridium piliforme 26-May-21 0 / 1 0 / 6 E q Mycoplasma pulmonis 26-May-21 0 / 1 0 / 6 E q Helicobacter spp.1 26-May-21 0 / 10 0 / 44 H q Salmonella spp. -
R Graphics Output
883 | Desulfovibrio vulgaris | DvMF_2825 298701 | Desulfovibrio | DA2_3337 1121434 | Halodesulfovibrio aestuarii | AULY01000007_gene1045 207559 | Desulfovibrio alaskensis | Dde_0991 935942 | Desulfonatronum lacustre | KI912608_gene2193 159290 | Desulfonatronum | JPIK01000018_gene1259 1121448 | Desulfovibrio gigas | DGI_0655 1121445 | Desulfovibrio desulfuricans | ATUZ01000018_gene2316 525146 | Desulfovibrio desulfuricans | Ddes_0159 665942 | Desulfovibrio | HMPREF1022_02168 457398 | Desulfovibrio | HMPREF0326_00453 363253 | Lawsonia intracellularis | LI0397 882 | Desulfovibrio vulgaris | DVU_0784 1121413 | Desulfonatronovibrio hydrogenovorans | JMKT01000008_gene1463 555779 | Desulfonatronospira thiodismutans | Dthio_PD0935 690850 | Desulfovibrio africanus | Desaf_1578 643562 | Pseudodesulfovibrio aespoeensis | Daes_3115 1322246 | Pseudodesulfovibrio piezophilus | BN4_12523 641491 | Desulfovibrio desulfuricans | DND132_2573 1121440 | Desulfovibrio aminophilus | AUMA01000002_gene2198 1121456 | Desulfovibrio longus | ATVA01000018_gene290 526222 | Desulfovibrio salexigens | Desal_3460 1121451 | Desulfovibrio hydrothermalis | DESAM_21057 1121447 | Desulfovibrio frigidus | JONL01000008_gene3531 1121441 | Desulfovibrio bastinii | AUCX01000006_gene918 1121439 | Desulfovibrio alkalitolerans | dsat_0220 941449 | Desulfovibrio | dsx2_0067 1307759 | Desulfovibrio | JOMJ01000003_gene2163 1121406 | Desulfocurvus vexinensis | JAEX01000012_gene687 1304872 | Desulfovibrio magneticus | JAGC01000003_gene2904 573370 | Desulfovibrio magneticus | DMR_04750 -

Analysis of Sequence Variation at Two Helicobacter Pylori Genetic Loci Potentially Involved in Virulence
W&M ScholarWorks Dissertations, Theses, and Masters Projects Theses, Dissertations, & Master Projects 2008 Analysis of Sequence Variation at Two Helicobacter pylori Genetic Loci Potentially involved in Virulence George Warren Liechti College of William & Mary - Arts & Sciences Follow this and additional works at: https://scholarworks.wm.edu/etd Part of the Microbiology Commons, and the Molecular Biology Commons Recommended Citation Liechti, George Warren, "Analysis of Sequence Variation at Two Helicobacter pylori Genetic Loci Potentially involved in Virulence" (2008). Dissertations, Theses, and Masters Projects. Paper 1539626867. https://dx.doi.org/doi:10.21220/s2-zrbg-b193 This Thesis is brought to you for free and open access by the Theses, Dissertations, & Master Projects at W&M ScholarWorks. It has been accepted for inclusion in Dissertations, Theses, and Masters Projects by an authorized administrator of W&M ScholarWorks. For more information, please contact [email protected]. Analysis of sequence variation atHelicobacter two pylori genetic loci potentially involved in virulence. George Warren Liechti Springfield, Virginia Bachelors of Science, College of William and Mary, 2003 A Thesis presented to the Graduate Faculty of the College of William and Mary in Candidacy for the Degree of Master of Science Department of Biology The College of William and Mary May, 2008 APPROVAL PAGE This Thesis is submitted in partial fulfillment of the requirements for the degree of Master of Science George Warren Liechti Approved by^the Cq , April, 2008 Committee Chair Associate Professor Mark Forsyth, Biology, The College of William and Mary r Professor Margaret Saha, Biology, The College of William and Mary Associate Professor George Gilchrist, Biology, The College of William and Mary / J / ABSTRACT PAGE Helicobacter pylori colonizes the gastric mucosa of nearly half the world’s population and is a well documented etiologic agent of peptic ulcer disease (PUD) and a significant risk factor for the development of gastric cancer.