Phase Behaviour and Miscibility Studies of Collagen/Silk Fibroin Macromolecular System in Dilute Solutions and Solid State
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

POLYMER STRUCTURE and CHARACTERIZATION Professor
POLYMER STRUCTURE AND CHARACTERIZATION Professor John A. Nairn Fall 2007 TABLE OF CONTENTS 1 INTRODUCTION 1 1.1 Definitions of Terms . 2 1.2 Course Goals . 5 2 POLYMER MOLECULAR WEIGHT 7 2.1 Introduction . 7 2.2 Number Average Molecular Weight . 9 2.3 Weight Average Molecular Weight . 10 2.4 Other Average Molecular Weights . 10 2.5 A Distribution of Molecular Weights . 11 2.6 Most Probable Molecular Weight Distribution . 12 3 MOLECULAR CONFORMATIONS 21 3.1 Introduction . 21 3.2 Nomenclature . 23 3.3 Property Calculation . 25 3.4 Freely-Jointed Chain . 27 3.4.1 Freely-Jointed Chain Analysis . 28 3.4.2 Comment on Freely-Jointed Chain . 34 3.5 Equivalent Freely Jointed Chain . 37 3.6 Vector Analysis of Polymer Conformations . 38 3.7 Freely-Rotating Chain . 41 3.8 Hindered Rotating Chain . 43 3.9 More Realistic Analysis . 45 3.10 Theta (Θ) Temperature . 47 3.11 Rotational Isomeric State Model . 48 4 RUBBER ELASTICITY 57 4.1 Introduction . 57 4.2 Historical Observations . 57 4.3 Thermodynamics . 60 4.4 Mechanical Properties . 62 4.5 Making Elastomers . 68 4.5.1 Diene Elastomers . 68 0 4.5.2 Nondiene Elastomers . 69 4.5.3 Thermoplastic Elastomers . 70 5 AMORPHOUS POLYMERS 73 5.1 Introduction . 73 5.2 The Glass Transition . 73 5.3 Free Volume Theory . 73 5.4 Physical Aging . 73 6 SEMICRYSTALLINE POLYMERS 75 6.1 Introduction . 75 6.2 Degree of Crystallization . 75 6.3 Structures . 75 Chapter 1 INTRODUCTION The topic of polymer structure and characterization covers molecular structure of polymer molecules, the arrangement of polymer molecules within a bulk polymer material, and techniques used to give information about structure or properties of polymers. -

Ideal Chain Conformations and Statistics
ENAS 606 : Polymer Physics Chinedum Osuji 01.24.2013:HO2 Ideal Chain Conformations and Statistics 1 Overview Consideration of the structure of macromolecules starts with a look at the details of chain level chemical details which can impact the conformations adopted by the polymer. In the case of saturated carbon ◦ backbones, while maintaining the desired Ci−1 − Ci − Ci+1 bond angle of 112 , the placement of the final carbon in the triad above can occur at any point along the circumference of a circle, defining a torsion angle '. We can readily recognize the energetic differences as a function this angle, U(') such that there are 3 minima - a deep minimum corresponding the the trans state, for which ' = 0 and energetically equivalent gauche− and gauche+ states at ' = ±120 degrees, as shown in Fig 1. Figure 1: Trans, gauche- and gauche+ configurations, and their energetic sates 1.1 Static Flexibility The static flexibility of the chain in equilibrium is determined by the difference between the levels of the energy minima corresponding the gauche and trans states, ∆. If ∆ < kT , the g+, g− and t states occur with similar probability, and so the chain can change direction and appears as a random coil. If ∆ takes on a larger value, then the t conformations will be enriched, so the chain will be rigid locally, but on larger length scales, the eventual occurrence of g+ and g− conformations imparts a random conformation. Overall, if we ignore details on some length scale smaller than lp, the persistence length, the polymer appears as a continuous flexible chain where 1 lp = l0 exp(∆/kT ) (1) where l0 is something like a monomer length. -

Native-Like Mean Structure in the Unfolded Ensemble of Small Proteins
B doi:10.1016/S0022-2836(02)00888-4 available online at http://www.idealibrary.com on w J. Mol. Biol. (2002) 323, 153–164 Native-like Mean Structure in the Unfolded Ensemble of Small Proteins Bojan Zagrovic1, Christopher D. Snow1, Siraj Khaliq2 Michael R. Shirts2 and Vijay S. Pande1,2* 1Biophysics Program The nature of the unfolded state plays a great role in our understanding of Stanford University, Stanford proteins. However, accurately studying the unfolded state with computer CA 94305-5080, USA simulation is difficult, due to its complexity and the great deal of sampling required. Using a supercluster of over 10,000 processors we 2Department of Chemistry have performed close to 800 ms of molecular dynamics simulation in Stanford University, Stanford atomistic detail of the folded and unfolded states of three polypeptides CA 94305-5080, USA from a range of structural classes: the all-alpha villin headpiece molecule, the beta hairpin tryptophan zipper, and a designed alpha-beta zinc finger mimic. A comparison between the folded and the unfolded ensembles reveals that, even though virtually none of the individual members of the unfolded ensemble exhibits native-like features, the mean unfolded structure (averaged over the entire unfolded ensemble) has a native-like geometry. This suggests several novel implications for protein folding and structure prediction as well as new interpretations for experiments which find structure in ensemble-averaged measurements. q 2002 Elsevier Science Ltd. All rights reserved Keywords: mean-structure hypothesis; unfolded state of proteins; *Corresponding author distributed computing; conformational averaging Introduction under folding conditions, with some notable exceptions.14 – 16 This is understandable since under Historically, the unfolded state of proteins has such conditions the unfolded state is an unstable, received significantly less attention than the folded fleeting species making any kind of quantitative state.1 The reasons for this are primarily its struc- experimental measurement very difficult. -
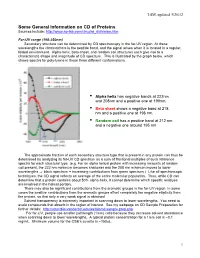
CD of Proteins Sources Include
LSM, updated 3/26/12 Some General Information on CD of Proteins Sources include: http://www.ap-lab.com/circular_dichroism.htm Far-UV range (190-250nm) Secondary structure can be determined by CD spectroscopy in the far-UV region. At these wavelengths the chromophore is the peptide bond, and the signal arises when it is located in a regular, folded environment. Alpha-helix, beta-sheet, and random coil structures each give rise to a characteristic shape and magnitude of CD spectrum. This is illustrated by the graph below, which shows spectra for poly-lysine in these three different conformations. • Alpha helix has negative bands at 222nm and 208nm and a positive one at 190nm. • Beta sheet shows a negative band at 218 nm and a positive one at 196 nm. • Random coil has a positive band at 212 nm and a negative one around 195 nm. The approximate fraction of each secondary structure type that is present in any protein can thus be determined by analyzing its far-UV CD spectrum as a sum of fractional multiples of such reference spectra for each structural type. (e.g. For an alpha helical protein with increasing amounts of random coil present, the 222 nm minimum becomes shallower and the 208 nm minimum moves to lower wavelengths ⇒ black spectrum + increasing contributions from green spectrum.) Like all spectroscopic techniques, the CD signal reflects an average of the entire molecular population. Thus, while CD can determine that a protein contains about 50% alpha-helix, it cannot determine which specific residues are involved in the helical portion. -

Large-Scale Analyses of Site-Specific Evolutionary Rates Across
G C A T T A C G G C A T genes Article Large-Scale Analyses of Site-Specific Evolutionary Rates across Eukaryote Proteomes Reveal Confounding Interactions between Intrinsic Disorder, Secondary Structure, and Functional Domains Joseph B. Ahrens, Jordon Rahaman and Jessica Siltberg-Liberles * Department of Biological Sciences, Florida International University, Miami, FL 33199, USA; [email protected] (J.B.A.); jraha001@fiu.edu (J.R.) * Correspondence: jliberle@fiu.edu; Tel.: +1-305-348-7508 Received: 1 October 2018; Accepted: 9 November 2018; Published: 14 November 2018 Abstract: Various structural and functional constraints govern the evolution of protein sequences. As a result, the relative rates of amino acid replacement among sites within a protein can vary significantly. Previous large-scale work on Metazoan (Animal) protein sequence alignments indicated that amino acid replacement rates are partially driven by a complex interaction among three factors: intrinsic disorder propensity; secondary structure; and functional domain involvement. Here, we use sequence-based predictors to evaluate the effects of these factors on site-specific sequence evolutionary rates within four eukaryotic lineages: Metazoans; Plants; Saccharomycete Fungi; and Alveolate Protists. Our results show broad, consistent trends across all four Eukaryote groups. In all four lineages, there is a significant increase in amino acid replacement rates when comparing: (i) disordered vs. ordered sites; (ii) random coil sites vs. sites in secondary structures; and (iii) inter-domain linker sites vs. sites in functional domains. Additionally, within Metazoans, Plants, and Saccharomycetes, there is a strong confounding interaction between intrinsic disorder and secondary structure—alignment sites exhibiting both high disorder propensity and involvement in secondary structures have very low average rates of sequence evolution. -

Effect of Starch on Property of Silk Fibroin/Keratin Blend Films
International Journal of GEOMATE, Dec., 2016, Vol. 11, Issue 28, pp.2870-2873 Special Issue on Science, Engineering and Environment, ISSN: 2186-2990, Japan EFFECT OF STARCH ON PROPERTY OF SILK FIBROIN/KERATIN BLEND FILMS Yaowalak Srisuwan, Ansaya Thonpho and Prasong Srihanam Faculty of Science, Mahasarakham University, Maha Sarakham 44150, Thailand ABSTRACT: This work was aimed to study the effect of starch on silk fibroin (SF)/keratin (K) blend films properties. The SF and K solutions were mixed with starch, homogeneously stirred and poured into polystyrene culture plates. The mixture solution was then dried in an oven at 40 °C for 3 days. The films were then investigated for their morphology, secondary structure and thermal properties by using scanning electron microscope (SEM), Fourier transform-infrared (FT-IR) spectrophotometer, Thermogravimetric analysis (TGA). The results found that each film had different patterns of surfaces depending on ratio used. The structure of almost films co-existed with random coil and α-helix structures which resulted to increase the flexibility and of film. The structure of the films changed to β-sheet after blending between SF and K according to H-bond formation and increased thermal stability of the films. This result indicated that starch helped to decrease the crystalline structure of the film which increased their flexibility. Keywords: Biopolymer, Morphology, Secondary structure, Thermal property 1. INTRODUCTION Silk fibroin (SF) solution was prepared by firstly boiling twice of B. mori cocoons in 0.5% (w/v) Silk is a natural fibrous protein produced from Na2CO3 solution at 90 °C for 30 min in each times, silkworm which had a unique characteristic. -

Protein Folding and Structure Prediction
Protein Folding and Structure Prediction A Statistician's View Ingo Ruczinski Department of Biostatistics, Johns Hopkins University Proteins Amino acids without peptide bonds. Amino acids with peptide bonds. ¡ Amino acids are the building blocks of proteins. Proteins Both figures show the same protein (the bacterial protein L). The right figure also highlights the secondary structure elements. Space Resolution limit of a light microscope Glucose Ribosome Red blood cell C−C bond Hemoglobin Bacterium 1 10 100 1000 10000 100000 1nm 1µm Distance [ A° ] Energy C−C bond Green Noncovalent bond light Glucose Thermal ATP 0.1 1 10 100 1000 Energy [ kcal/mol ] Non-Bonding Interactions Amino acids of a protein are joined by covalent bonding interactions. The polypep- tide is folded in three dimension by non-bonding interactions. These interactions, which can easily be disrupted by extreme pH, temperature, pressure, and denatu- rants, are: Electrostatic Interactions (5 kcal/mol) Hydrogen-bond Interactions (3-7 kcal/mol) Van Der Waals Interactions (1 kcal/mol) ¡ Hydrophobic Interactions ( 10 kcal/mol) The total inter-atomic force acting between two atoms is the sum of all the forces they exert on each other. Energy Profile Transition State Denatured State Native State Radius of Gyration of Denatured Proteins Do chemically denatured proteins behave as random coils? The radius of gyration Rg of a protein is defined as the root mean square dis- tance from each atom of the protein to their centroid. For an ideal (infinitely thin) random-coil chain in a solvent, the average radius 0.5 ¡ of gyration of a random coil is a simple function of its length n: Rg n For an excluded volume polymer (a polymer with non-zero thickness and non- trivial interactions between monomers) in a solvent, the average radius of gyra- 0.588 tion, we have Rg n (Flory 1953). -

Programming Function Into Mechanical Forms by Directed Assembly of Silk
Programming function into mechanical forms by SEE COMMENTARY directed assembly of silk bulk materials Benedetto Marellia,1, Nereus Patela, Thomas Duggana, Giovanni Perottoa, Elijah Shirmana, Chunmei Lia, David L. Kaplana,b, and Fiorenzo G. Omenettoa,c,d,2 aSilklab, Department of Biomedical Engineering, Tufts University, Medford, MA 02155; bDepartment of Chemical Engineering, Tufts University, Medford, MA 02155; cDepartment of Electrical Engineering, Tufts University, Medford, MA 02155; and dDepartment of Physics, Tufts University, Medford, MA 02155 Edited by David A. Weitz, Harvard University, Cambridge, MA, and approved November 21, 2016 (received for review July 23, 2016) We report simple, water-based fabrication methods based on particular, three different models have been proposed for silk protein self-assembly to generate 3D silk fibroin bulk materials assembly, which have been supported by diverse experimental that can be easily hybridized with water-soluble molecules to evidences obtained from Bombyx mori silk, spider silk proteins, obtain multiple solid formats with predesigned functions. Con- regenerated silk fibroin, and recombinantly produced spider trolling self-assembly leads to robust, machinable formats that silk proteins at different concentrations (9–18). A recent review exhibit thermoplastic behavior consenting material reshaping at covers in detail these models (19). (i) The first mechanism the nanoscale, microscale, and macroscale. We illustrate the involves the formation of micellar nanostructures that fuse versatility of the approach by realizing demonstrator devices together via coalescence, forming microscopic globular struc- where large silk monoliths can be generated, polished, and tures that, in the presence of shear stress, elongate and fuse reshaped into functional mechanical components that can be together (9). -

Silk-Based Biomaterials for Tissue Engineering
C H A P T E R 1 Silk-based Biomaterials for Tissue Engineering V. Kearns, A.C. MacIntosh, A. Crawford and P.V. Hatton* Summary ilks are fibrous proteins, which are spun by a variety of species including silkworms and spiders. Silks have common structural components and have a hierarchical structure. Silkworm silk must be S degummed for biomedical applications in order to remove the immunogenic sericin coating. It may subsequently be processed into a variety of forms, often via the formation of a fibroin solution, including films, fibres and sponges, and used in combination with other materials such as gelatine and hydroxyapatite. Spider silks do not have a sericin coating and may be used in natural fibre form or processed via formation of a spidroin solution. Both silkworm and spider silks have been reported to support attachment and proliferation of a variety of cell types. Silks have subsequently been investigated for use in tissue engineering. This chapter provides a general overview of silk biomaterials, discussing their processing, biocompatibility and degradation behaviour and paying particular attention to their applications in tissue engineering. KEYWORDS: Silk; Fibroin; Spidroin; Tissue engineering. *Correspondence to: Professor Paul V Hatton, Centre for Biomaterials & Tissue Engineering, School of Clinical Dentistry, University of Sheffield, Claremont Crescent, Sheffield, S10 2TA, UK. Email: [email protected] Tel: +44 (114) 271 7938 Fax: +44 (114) 279 7050. Topics in Tissue Engineering, Vol. 4. Eds. N Ashammakhi, R Reis, & F Chiellini © 2008. Kearns et al. Silk Biomaterials 1. INTRODUCTION Silks are fibrous proteins, which are spun into fibres by a variety of insects and spiders [1]. -

The Physiological Accumulation of Mutant Fibroin Light Chains Induces
Journal of Insect Biotechnology and Sericology 86, 105-112 (2017) The physiological accumulation of mutant fibroin light chains induces an unfolded protein response in the posterior silk gland of the Sericin cocoon Nd-sD strain of silkworm Bombyx mori Tadashi Takahashi1, Masao Miyazaki1, Shin-ichiro Kidou2, Yoshiki Matsui1, Ying An1, Taku Ozaki3, Koichi Suzuki1** and Tetsuro Yamashita1* 1 Faculty of Agriculture, Iwate University, Ueda, Morioka, Iwate 020-8550, Japan 2 Graduate School of Natural Sciences, Nagoya City University, Mizuho-cho, Mizuho-ku, Nagoya 467-8501, Japan 3 Faculty of Science and Engineering, Iwate University, Ueda, Morioka, Iwate 020-8550, Japan (Received May 15, 2017; Accepted July 4, 2017) Fibroin, a major component of silk fiber, is composed of light (L) chains, heavy chains, and fhx/P25 in the silk- worm Bombyx mori. Sericin cocoon (Nd-sD) is a silkworm strain expressing a mutant fibroin L chain that is accu- mulated in the endoplasmic reticulum (ER) of posterior silk glands (PSGs). However, little is known about the effects of accumulation in PSGs in Nd-sD strain. We compared the PSG gene expression profiles of 5th-instar larvae of Nd-sD strain and the fibroin-producing normal strain. cDNA representational difference analysis identi- fied candidate genes whose expression levels were higher in Nd-sD strain than in normal strain. We focused on heat shock proteins and cathepsin B, a major lysosomal protease. We confirmed upregulation of BiP(GRP78), Hsp20.8, and cathepsin B at the transcriptional and/or translational levels. These results suggest that the accu- mulation of mutant L chain induces the unfolded protein response in PSGs of Nd-sD strain, which will be a valu- able tool for protein quality-control studies of silk grand. -

Helix-Coil Conformational Change Accompanied by Anisotropic–Isotropic Transition
Polymer Journal, Vol.33, No. 11, pp 898—901 (2001) NOTES Helix-Coil Conformational Change Accompanied by Anisotropic–Isotropic Transition † Ning LIU, Jiaping LIN, Tao C HEN, Jianding CHEN, Dafei ZHOU, and Le LI Department of Polymer Science and Engineering, East China University of Science and Technology, Shanghai 200237, P. R. China (Received November 22, 2000; Accepted July 1, 2001) KEY WORDS Helix-Coil Transition / Liquid Crystal Polymer / Polypeptide / Anisotropic– Isotropic Transition / Polypeptides, particularly poly(γ-benzyl-L- limited study of this acid induced anisotropic–isotropic glutamate) (PBLG) and other glutamic acid ester transition has been reported.13, 14 The precise molecular polymers, have the ability to exist in α-helix, a well de- mechanism of the transition remains to be determined. fined chain conformation of long chain order and retain In the present work, the texture changes in the acid- such a structure in numerous organic solvents that sup- induced anisotropic–isotropic transition were studied port intramolecular hydrogen bonding.1, 2 If PBLG and by microscope observations and the corresponding related polypeptides are dissolved in a binary solvent molecular conformational changes were followed by IR mixture containing a non-helicogenic component such measurements. The effect of the polymer concentra- as dichloroacetic acid (DCA) or trifluoroacetic acid tion was studied. According to the information gained (TFA), they can undergo a conformational change from through the experiment, the molecular mechanism with the α-helix to random coil form due to the changes respect to the acid-induced helix-coil conformational in solvent composition, temperature or both.3, 4 The change was suggested. -

A Review of Structure Construction of Silk Fibroin Biomaterials from Single Structures to Multi-Level Structures
International Journal of Molecular Sciences Review A Review of Structure Construction of Silk Fibroin Biomaterials from Single Structures to Multi-Level Structures Yu Qi 1, Hui Wang 1,*, Kai Wei 1, Ya Yang 1, Ru-Yue Zheng 1, Ick Soo Kim 2 and Ke-Qin Zhang 1,* 1 National Engineering Laboratory for Modern Silk, College of Textile and Clothing Engineering, Soochow University, Suzhou 215123, China; [email protected] (Y.Q.); [email protected] (K.W.); [email protected] (Y.Y.); [email protected] (R.-Y.Z.) 2 Nano Fusion Technology Research Lab, Interdisciplinary Cluster for Cutting Edge Research (ICCER), Division of Frontier Fibers, Institute for Fiber Engineering (IFES), Shinshu University, Ueda, Nagano 386 8567, Japan; [email protected] * Correspondence: [email protected] (H.W.); [email protected] (K.-Q.Z.) Academic Editors: John G. Hardy and Chris Holland Received: 24 November 2016; Accepted: 11 January 2017; Published: 3 March 2017 Abstract: The biological performance of artificial biomaterials is closely related to their structure characteristics. Cell adhesion, migration, proliferation, and differentiation are all strongly affected by the different scale structures of biomaterials. Silk fibroin (SF), extracted mainly from silkworms, has become a popular biomaterial due to its excellent biocompatibility, exceptional mechanical properties, tunable degradation, ease of processing, and sufficient supply. As a material with excellent processability, SF can be processed into various forms with different structures, including particulate, fiber, film, and three-dimensional (3D) porous scaffolds. This review discusses and summarizes the various constructions of SF-based materials, from single structures to multi-level structures, and their applications.