HELQ Is a Dual Function DSB Repair Enzyme Modulated by RPA and RAD51
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
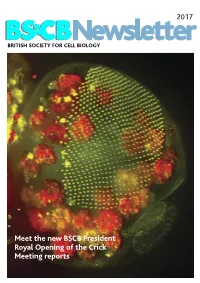
BSCB Newsletter 2017D
2017 BSCB Newsletter BRITISH SOCIETY FOR CELL BIOLOGY Meet the new BSCB President Royal Opening of the Crick Meeting reports 2017 CONTENTS BSCB Newsletter News 2 Book reviews 7 Features 8 Meeting Reports 24 Summer students 30 Society Business 33 Editorial Welcome to the 2017 BSCB newsletter. After several meeting hosted several well received events for our Front cover: years of excellent service, Kate Nobes has stepped PhD and Postdoc members, which we discuss on The head of a Drosophila pupa. The developing down and handed the reins over to me. I’ve enjoyed page 5. Our PhD and Postdoc reps are working hard compound eye (green) is putting together this years’ newsletter. It’s been great to make the event bigger and better for next year! The composed of several hundred simple units called ommatidia to hear what our members have been up to, and I social events were well attended including the now arranged in an extremely hope you will enjoy reading it. infamous annual “Pub Quiz” and disco after the regular array. The giant conference dinner. Members will be relieved to know polyploidy cells of the fat body (red), the fly equivalent of the The 2016 BSCB/DB spring meeting, organised by our we aren’t including any photos from that here. mammalian liver and adipose committee members Buzz Baum (UCL), Silke tissue, occupy a big area of the Robatzek and Steve Royle, had a particular focus on In this issue, we highlight the great work the BSCB head. Cells and Tissue Architecture, Growth & Cell Division, has been doing to engage young scientists. -

Gagna A. & Ch. Van Heck
GAGNA A. & CH. VAN HECK PRIZE 2015 18 NOVEMBER 2015 Contact: Mr Bruno MORAUX F.R.S.-FNRS [email protected] 02/504.92.40 GAGNA A. & CH. VAN HECK PRIZE The triennial and international « Gagna A. & Ch. Van Heck Prize », amounting to 75.000 €, rewards a scientist or a Medical Doctor, in recognition of a research work which has contributed to the treatment of a disease currently incurable, or which has raised hopes for curing the disease. This Prize was first granted in 2003 and constitutes one of the most prestigious Belgian Prize in biomedical science. The Prize is awarded to: Stephen P. JACKSON Ph.D. in Molecular Biology, University of Edinburgh (UK) Head of Cancer Research UK Laboratories, The Gurdon Institute, University of Cambridge (UK) Professor of Biology, University of Cambridge (UK) Identification of key mechanisms for the detection, signaling and repair of DNA damages. Professor Stephen Jackson’s pioneering academic research has provided us with many of the key principles by which cells detect DNA damage, signal its presence and mediate its repair. His discoveries have had particularly major impacts on our understanding of how human cells deal with the most toxic of all DNA damages: the DNA double-strand break (DSB). In addition to establishing some of the most important foundations of present-day DNA repair research, Jackson’s work has also shown us how cellular dysfunction and disease arise when DNA repair or DNA-damage signaling goes awry. Additionally, his work has led to the development of an innovative new approach to cancer therapy that kills cancer cells whose ability to repair DNA damage has been compromised. -
Life Changing Science
LIFE CHANGING SCIENCE The Francis Crick Institute Annual Review 2017/18 AN INSTITUTE FOR DISCOVERY Our commitment to excellence, our emphasis on multidisciplinary research, our focus on young and emerging talent and our novel ways of partnership working are some of the factors that set the Crick apart. Front cover Vaccinia virus infection (green) disrupts a layer of epithelial cells (red/blue). Courtesy of Michael Way, Group Leader at the Crick. INTRODUCTION 2 Who we are Our year at a glance 2 Introduction by Paul Nurse 4 The Francis Crick Institute is a biomedical Progress against our strategy 6 discovery institute dedicated to understanding the RESEARCH HIGHLIGHTS 10 Cancer-causing mutation fundamental biology underlying health and disease. suppresses immune system 11 Our work is helping to build an understanding of Predicting lung cancer’s return 12 New understanding of human why disease develops and to translate discoveries embryo development 14 Chemical attraction could improve into new ways to prevent, diagnose and treat cancer immunotherapy 16 illnesses such as cancer, heart disease, stroke, Genes linked to malaria parasites’ persistence 17 infections and neurodegenerative diseases. Architecture of our ‘second brain’ 18 Cause of infertility side-stepped in mice 19 Mechanism for spinal cord development discovered 20 A new layer of complexity in embryo development 21 Two DNAs wedded with this ring 22 Unravelling how DNA gets copied 23 Telomerase’s dark side discovered 24 REVIEW OF THE YEAR 26 New group leaders arrive 27 Joined-up thinking 30 Focusing on the molecules of life 32 CryoEM at the Crick 34 Bringing academia and industry closer together 36 The people making research happen 38 Patterns in art and science 40 Rewarding research 42 Appointments 43 Supporting new discoveries 44 Our vision What’s inside Our vision is to be a world- We bring together outstanding scientists Science feature 32 leading multidisciplinary from all disciplines and carry out research Sophisticated microscopy is being biomedical research institute. -

Doctoral Fellowships for Clinicians 2018
DOCTORAL FELLOWSHIPS FOR CLINICIANS 2018 Applications are invited for Crick–Cancer Research UK and Crick doctoral fellowships for clinicians at the Francis Crick Institute (the Crick). These fellowships provide an opportunity for outstanding clinicians to perform innovative biomedical discovery research for three years while following the Crick’s clinical PhD programme. The Crick carries out multidisciplinary research Fellows will be based at the Crick, supported by teams dedicated to unravelling the scientific mechanisms of comprising a Crick research group leader, at least one living organisms, helping us to understand why disease supervisor from one of Crick’s five clinical PhD programme develops and find new ways to prevent, diagnose and partners (Barts Cancer Institute/Queen Mary University treat illnesses. One of the Crick’s aims is to provide an of London, Imperial College London, The Institute of environment to advance the understanding of cancer at Cancer Research, King’s College London and UCL) and at the most fundamental level and to speed application of least one clinician. this knowledge for therapeutic and diagnostic patient benefit. Crick-Cancer Research UK projects • Cancer outcome in standard and immune checkpoint blockade therapies. Dinis Calado (Crick), Jude Fitzgibbon (Barts Cancer Institute/QMUL) and Peter Johnson (University of Southampton) • Investigation of the mechanisms by which RAS mutant lung cancer evades the immune system. Julian Downward (Crick), Alan Melcher (ICR) and Sanjay Popat (ICR, Royal Marsden) • Genomic assessment of DNA damage in skin cancers. Nicholas Luscombe (Crick), Fiona Watt and Magnus Lynch (King’s College London) • Identification of novel targets to inhibit autophagy in advanced pancreatic cancer. -

Communicating Biochemistry: Meetings and Events
© The Authors. Volume compilation © 2011 Portland Press Limited Chapter 3 Communicating Biochemistry: Meetings and Events Ian Dransfield and Brian Beechey Scientific conferences organized by the Biochemical Society represent a key facet of activity throughout the Society’s history and remain central to the present mission of promoting the advancement of molecular biosciences. Importantly, scientific conferences are an important means of communicating research findings, establishing collaborations and, critically, a means of cementing the community of biochemical scientists together. However, in the past 25 years, we have seen major changes to the way in which science is communicated and also in the way that scientists interact and establish collabo- rations. For example, the ability to show videos, “fly through” molecular structures or show time-lapse or real-time movies of molecular events within cells has had a very positive impact on conveying difficult concepts in presentations. However, increased pressures on researchers to obtain/maintain funding can mean that there is a general reluctance to present novel, unpublished data. In addition, the development of email and electronic access to scientific journals has dramatically altered the potential for communi- cation and accessibility of information, perhaps reducing the necessity of attending meetings to make new contacts and to hear exciting new science. The Biochemical Society has responded to these challenges by progressive development of the meetings format to better match the -

Summary • Editorial June 2019 • 31St
Summary • Editorial June 2019 • 31st Pezcoller Symposium - Program - Abstracts of oral presentations - Abstracts of posters • Call for nomination 2020 Pezcoller Foundation-AACR International Award for Extraordinary Achievement in Cancer Research • Call for nomination Scholar-In-Training Awards News from the Year 29 – No. 52 Pezcoller Foundation World June 2019 Picture on front page: 2019 Pezcoller Foundation-AACR International Award for Extraordinary Achievement in Cancer Research winner: Prof. Alberto Mantovani June 2019 Editorial Integrative Biology) of the University of Trento. Editorial In the Award Ceremony, President Galligioni highlighted the main points of Mantovani’s re- searches and discoveries. “Dr. Mantovani is an eminent physician-scien- tist, he said, a leader in the field of tumor im- munology for decades. Presently Scientific Di- rector at the Istituto Clinico Humanitas and Full Professor of General Pathology at the Humanitas University, Mantovani was for many years Head of the Department of Immunology and Cell Biol- ogy, Istituto di Ricerche Farmacologiche “Mario Negri”, Milan, Italy. Furthermore he holds the Chair of Inflammation and Therapeutic Innova- tion, at the Queen Mary University, London, UK. By identifying macrophages in tumors as corrupt- ed policemen promoting cancer progression, and by discovering relevant genes and functions, Al- berto Mantovani highlighted the role of inflam- mation and immunity in the tumor microenvi- First of all we are reporting with a great pleas- ronment, a paradigm shift fundamental for the ure that the recipient of 2019 Pezcoller Foun- development of tumor immunology and immu- dation – AACR International Award for Extraor- notherapy. In addition, his researches includ- dinary Achievement in Cancer research is Prof. -

Cancer Research UK Dr Maximilian Martin, Global Head of Philanthropy, Bank Lombard Odier & Co Lt
Cancer research funding—Navigating a competitive environment The role of collaboration and innovation 4 April 2019 Chair Miranda Brawn, Senior Business, Legal and Diversity Executive and Founder of The Miranda Brawn Diversity Leadership Foundation Panel Jonathan Tobin, Investment Director, Arix Biosciences Tony Hickson, Chief Business Officer, Cancer Research UK Dr Maximilian Martin, Global Head of Philanthropy, Bank Lombard Odier & Co Lt Cancer Research UK April 2019 TRANSLATION (DRUGS): IMPACT ON PATIENTS MULTIPLE COMMERCIALISATION APPROACHES Alliances 6 major partner alliances in our Therapeutic Discovery Labs Licences 120 existing licences Collaborations 21 collaborations completed as of 2018 Spin-outs We have been involved in the formation and development of over 30 spin-out companies 6 MULTIPLE COMMERCIALISATION APPROACHES Alliances 6 major partner alliances in our Therapeutic Discovery Labs Licences 120 existing licences Collaborations 21 collaborations completed as of 2018 Spin-outs We have been involved in the formation and development of over 30 spin-out companies 7 THE ARTIOS DEVELOPMENT STORY Nov-16 May-16 Sept-16 Dr Simon Boulton Sept-17 Feb-18 Aug-18 Seed funding Series A appointed as VP Dr Graeme Smith Pierre Legault Series B through CRUK fundraisin Science Strategy appointed as Chief appointed as fundraising of and SV Health g of Scientific Officer Chairman of BoD $84m Partners $33.2m 2016 2017 2018 Sept-16 Sept-17 May-16 Artios in-licenses Mar-18 Artios wins Biotech & CRUK and SV potential first-in- Artios exercises option -

The End Protection Problem—An Unexpected Twist in the Tail
Downloaded from genesdev.cshlp.org on October 3, 2021 - Published by Cold Spring Harbor Laboratory Press PERSPECTIVE The end protection problem—an unexpected twist in the tail Phil Ruis and Simon J. Boulton The Francis Crick Institute, London NW1 1AT, United Kingdom In this perspective, we introduce shelterin and the and cell death, the question of how shelterin—and, espe- mechanisms of ATM activation and NHEJ at telomeres, cially, TRF2—protects chromosome ends from DNA re- before discussing the following questions: How are t- pair activities has been paramount. A succession of loops proposed to protect chromosome ends and what is papers followed, showing that loss of TRF2 leads to activa- the evidence for this model? Can other models explain tion of the DNA double strand break (DSB) kinase ATM how TRF2 mediates end protection? Could t-loops be and classical nonhomologous end joining (NHEJ) at telo- pathological structures? How is end protection achieved meres. The question of how TRF2 protects chromosome in pluripotent cells? What do the insights into telomere ends has thus focused on how TRF2 prevents the activa- end protection in pluripotent cells mean for the t-loop tion of these dual pathways—ATM and NHEJ—at telo- model of end protection? Why might different cell states meres. Both pathways rely on the binding of DSB have evolved different mechanisms of end protection? sensors (MRE11/RAD50/NBS1 [MRN] and KU70/80, re- Finally, we offer support for an updated t-loop model of spectively) to the same substrate (DNA ends), suggesting end protection, suggesting that the data is supportive of that one possible way TRF2 could inhibit these pathways a critical role for t-loops in protecting chromosome ends would be to hide these DSB ends. -

Illustrated Holocene Era Timeline
Illustrated Holocene Era Timeline: Human Achievements, Advancements, Innovations, and Understanding in Science using EMILIANI’s HE calendar Ruthie S. Premack Author / Compiler Technical Advisor Paul Premack Phone Edition 12,019 HE I dedicate this book To: the wonderful man who is my husband Paul Premack, our adult children Tiffany and Benjamin, his wife Kira, my mother Jo Ann Simons Stier for their love, brains, attention to detail, laughter, and thoughtfulness and to my dad Herb Stier; To: CESARE EMILIANI, who first had the idea for the Holocene Era (HE) calendar; and To: any human who can open their mind to seeing the (HE) flow of human accomplishment and to being enchanted, shocked, disappointed, or amazed by the wonders and realities of science. © 2019 (12,019 HE), Ruthie S. Premack, all rights reserved. Any portion, but not the entirety, of this book may be reproduced without permission from the author so long as this book is cited as the source and entries are not altered. Some text in this Timeline is from or based upon entries in Wikipedia. Use is permitted because we have provided credit to the authors by including a URL to the page or pages used. All entries based upon Wikipedia materials is released under CC BY-SA: https://creativecommons.org/licenses/by-sa/3.0/ Table of Contents ABOUT THE HE CALENDAR AND FORMATTING ............................... 7 AUTHOR / COMPILER’S PREFACE ........................................................ 10 CHAPTER ONE APPETIZERS ............................................................. 12 CHAPTER TWO BEFORE THE HOLOCENE ERA: THE BIG BANG TO THE STONE AGE .............................................. 45 CHAPTER THREE 1 HE: BEGINNING OF THE HOLOCENE ERA .................................................................................... -

ANNUAL REPORT 2014 2 Centre for Cancer Research & Cell Biology | Annual Report | 2014 CONTENTS
ANNUAL REPORT 2014 2 Centre for Cancer Research & Cell Biology | Annual Report | 2014 CONTENTS Director’s Introduction 4 Education and Training 28 Overview of Centre Strategy 5 Clinical Academic Training Programme 29 Centre Programmes 6 Post Doctoral Programme 30 Centre Life 8 Seminar Programme 32 Research Awards 9 Postgraduate Programme 34 Undergraduate Programme 37 Focus Groups 10 Advanced Radiotherapy 11 Public Engagement Activities 38 Breast 12 Blood Cancers 13 Staff Listing 41 Genito-Urinary/Prostate 14 New Appointments 42 Gastro-Intestinal 16 Current Staff 46 Brain Tumours 18 Genomics 19 Major Sources of Funding 49 Ovarian 20 Funding Bodies 50 Research Grants Awarded 51 Enabling Technologies 21 Drug Discovery 22 Publications 54 Molecular Pathology and Biobanking 24 Translational Bioinformatics and Imaging 26 Acknowledgements 63 Northern Ireland Cancer Trials Centre 27 and Network Centre for Cancer Research & Cell Biology | Annual Report | 2014 3 DIRECTOR’S INTRODUCTION DAVID WAUGH, Director The Centre for Cancer Research and Cell Biology (CCRCB) is the beating heart of a comprehensive cancer research programme in Belfast, focused on accelerating the delivery of scientific discovery to clinical application. CCRCB is the research hub of the Belfast Cancer Research UK Centre and the Belfast Experimental Cancer Medicine Centre (ECMC), operating in partnership with the Belfast Health and Social Care Trust. Our integrated clinical and scientific programmes address clinically unmet needs. Our principal objective is to promote evidence-based, innovative clinical trials in order to underpin improved patient outcomes in high incidence solid tumours of Gastro-Intestinal, Prostatic, Breast and Ovarian origin, and in specific Blood Cancers. Our unifying research theme is to develop translational outputs, in the form of biomarkers and/or novel therapeutic strategies that enable our Centre to be at the forefront of personalized cancer medicine in these prevalent diseases. -

2012 Fellowship Election Results
6/14/13 Academy of Medical Sciences - September 2011 - Newsletter 2012 Fellowship election results 46 of the UK's leading medical researchers have been recognised for excellence in medical science with their election to the Academy Fellowship. These distinguished scientists bring the total number of Academy Fellows to 1061. We offer the new Fellows our warmest congratulations on their election and welcome them to the Academy. Details of these new Fellows can be viewed on the Academy website where you can also download nomination forms for the 2013 election. The new Fellows elected in 2011 Following a drive to encourage nominations of female biomedical researchers, fifteen (33%) of these new Fellows are women a significant improvement on the number elected in previous years and more than double the percentage of female Professors in the biosciences. For further information contact [email protected] Fellows elected in 2012 Professor Christopher Abell FMedSci, Professor of Biological Chemistry, University of Cambridge Professor Dario Alessi FMedSci, Director of MRC Protein Phosphorylation and Ubiquitylation Unit, University of Dundee Professor Deborah Ashby OBE FMedSci, Professor of Medical Statistics and Clinical Trials, Co-Director of Imperial Clinical Trials Unit, Imperial College London Professor Tipu Aziz FMedSci, Professor of Neurosurgery, University of Oxford Professor Denis Azzopardi FMedSci, Professor of Neonatal Medicine, Centre for the Developing Brain, Imperial College London and Kings College London Professor Julian -

Annual Report & Financial Statements
Annual Report & Financial Statements for the year ended 31st December 2019 Action for A-T - Charity registered number 1145303 www.ActionforAT.org 1 Legal And Reference Information Patrons of Cambridge Professor Grant Stewart, Professor of Cancer Genetics, Jonny Wilkinson CBE School of Cancer Sciences, University of Birmingham Roger Black MBE Dr Alastair Wilkins, Reader in Neurology, University of Bristol Jeremy Guscott MBE and Consultant Neurologist at Frenchay Hospital, Bristol. Simon Shaw MBE Rachel Morris MBE Professor Matthew Wood, Professor of Neuroscience and Deputy Head of the Medical Sciences Division at the Naga Munchetty University of Oxford Leon Haslam Patient Representatives Chief Executive Mr Amandeep Sharma Sean Kelly Mrs Clare Gallagher Trustees at the date of approval Charity Registration Number Toby Read (Chairman) United Kingdom Registered Charity number: 1145303 Emily Read William Rowberry (Treasurer) Tomos Shillingford Registered Office Maria Leonard Aruna House Chris Askew 2 Kings Road Dr Mark Toms Haslemere Joseph Frost Surrey GU27 2QA Resigned during the year Samantha Cornell 1st December 2019 Operational Address Action for A-T Appointed during the year or since year end Carlton House 19 West Street Joseph Frost 1st January 2020 Epsom Surrey KT18 7RL Research Advisory Committee Dr Mark Toms, Chairman Professional Advisers Dr Kathryn Johnson, Scientific Advisor, Consultant Neonatologist and Research Lead Solicitors Bingham McCutchen LLP Professor David Attwell, Jodrell Professor of Physiology, Department of Neuroscience,