Supplementary Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
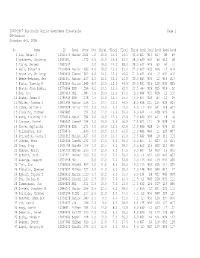
2006-2007 Wisconsin Junior Open/Open Crosstable Page 1 UW-Oshkosh November 4-5, 2006
2006-2007 Wisconsin Junior Open/Open Crosstable Page 1 UW-Oshkosh November 4-5, 2006 No. Name ID Team Rate Pts TBrk1 TBrk2 TBrk3 TBrk4 Rnd1 Rnd2 Rnd3 Rnd4 Rnd5 1 Luo, Brian J 12910173 Madisn 2024 5.0 16.0 15.0 54.0 32.0 W27 W13 W17 W9 W3 2 Mckinney, Christop 12934651 1772 4.5 16.0 14.5 52.0 28.0 W30 W33 W6 W10 D8 3 Yusim, Sergey 12852542 4.0 19.0 14.0 58.0 28.0 W12 W29 W5 W4 L1 4 Bell, Samuel W 21014944 Madisn 1663 4.0 15.0 13.0 51.0 22.0 W22 W35 W16 L3 W14 5 Brown Jr, Christop 12849430 GlenNi 1501 4.0 14.5 12.0 46.0 21.0 W31 W34 L3 W21 W16 6 Webne-Behrman, Ger 12901591 Madisn 1437 4.0 14.5 12.0 43.5 20.0 W43 W36 L2 W19 W13 7 Hintz, Timothy M 12752556 Marion 1545 4.0 13.5 12.0 44.5 20.0 W37 W19 L10 W33 W25 8 Bowen, John Armbru 12779264 USM 1596 4.0 13.5 11.5 42.0 22.5 -H- W26 W15 W14 D2 9 Her, Sou 12805616 MSL 1687 3.5 16.5 12.5 51.0 19.5 W44 W21 W18 L1 D10 10 Bowen, James H 12786949 USM 1278 3.5 14.0 12.5 45.0 15.5 W53 W49 W7 L2 D9 11 Miller, Joshua D 12871494 Marion 1516 3.5 13.5 10.0 44.5 18.0 W38 D15 L14 W24 W29 12 Dumke, Nathan C 12849428 Marion 1227 3.5 13.5 9.0 43.0 16.0 L3 W41 W34 D18 W27 13 Clausing, Michael 12864685 1254 3.0 17.0 10.0 50.0 16.0 W24 L1 W38 W20 L6 14 Wang, Xiaoming Tim 12976014 MadJf 1254 3.0 16.5 12.0 47.5 17.0 W40 W20 W11 L8 L4 15 Lancour, Daniel 12859342 JanesP 1158 3.0 16.0 9.5 46.5 17.5 W23 D11 L8 W28 D18 16 Ghose, Saptarshi 12870578 USM 1271 3.0 14.5 11.0 42.5 13.0 W45 W28 L4 W39 L5 17 Gallenberg, Bob 12779870 1436 3.0 13.5 10.0 41.5 11.0 W41 W46 L1 L25 W37 18 Petrashek, Casey D 12911815 Madisn 1427 3.0 13.0 -

Heater Element Specifications Bulletin Number 592
Technical Data Heater Element Specifications Bulletin Number 592 Topic Page Description 2 Heater Element Selection Procedure 2 Index to Heater Element Selection Tables 5 Heater Element Selection Tables 6 Additional Resources These documents contain additional information concerning related products from Rockwell Automation. Resource Description Industrial Automation Wiring and Grounding Guidelines, publication 1770-4.1 Provides general guidelines for installing a Rockwell Automation industrial system. Product Certifications website, http://www.ab.com Provides declarations of conformity, certificates, and other certification details. You can view or download publications at http://www.rockwellautomation.com/literature/. To order paper copies of technical documentation, contact your local Allen-Bradley distributor or Rockwell Automation sales representative. For Application on Bulletin 100/500/609/1200 Line Starters Heater Element Specifications Eutectic Alloy Overload Relay Heater Elements Type J — CLASS 10 Type P — CLASS 20 (Bul. 600 ONLY) Type W — CLASS 20 Type WL — CLASS 30 Note: Heater Element Type W/WL does not currently meet the material Type W Heater Elements restrictions related to EU ROHS Description The following is for motors rated for Continuous Duty: For motors with marked service factor of not less than 1.15, or Overload Relay Class Designation motors with a marked temperature rise not over +40 °C United States Industry Standards (NEMA ICS 2 Part 4) designate an (+104 °F), apply application rules 1 through 3. Apply application overload relay by a class number indicating the maximum time in rules 2 and 3 when the temperature difference does not exceed seconds at which it will trip when carrying a current equal to 600 +10 °C (+18 °F). -

K-12 Individual No. Name Team Gr Rate Pts Tbrk1 Tbrk2 Tbrk3 Tbrk4
K-12 Individual No. Name Team Gr Rate Pts TBrk1 TBrk2 TBrk3 TBrk4 Rnd1 Rnd2 Rnd3 Rnd4 Rnd5 Rnd6 1 Chakraborty, Dipro 11 2299 5.5 21 24 43 20.5 W27 W12 W5 W32 W8 D3 State Champion, AZ Denker Representative 2 Yim, Tony Sung BASISS 8 2135 5 20.5 23.5 38.5 17.5 W24 W10 D3 D16 W11 W9 3 Aletheia-Zomlefer, Soren CHANPR 11 1961 5 20 23 35.5 18.5 W25 W26 D2 W40 W15 D1 4 Desmarais, Nicholas Eduard NOTRED 10 1917 5 18 20 33 18 W39 W23 W18 L15 W10 W8 5 Wong, Kinsleigh Phillip CFHS 10 1992 4.5 20 20 24.5 15 -X- W17 L1 W26 D7 W15 6 Todd, Bryce BASISC 10 1923 4.5 17 19 26.5 14.5 W38 D18 L9 W23 W21 W16 7 Chaliki, Kalyan DSMTHS 9 1726 4.5 17 18.5 26 15 W46 L16 W28 W22 D5 W17 8 Li, Bohan UHS 9 2048 4 22 25 29 18 W30 W11 W45 W9 L1 L4 9 Mittal, Rohan CFHS 9 1916 4 19.5 20.5 23 17 W47 W22 W6 L8 W20 L2 10 Pennock, Joshua CFHS 10 1682 4 19 22 24 14 W31 L2 W25 W21 L4 W29 11 Aradhyula, Sumhith CFHS 9 1631 4 18 20 22 14 W41 L8 W38 W13 L2 W19 12 Johnston, Nicolas Godfrey CFHS 9 1803 4 18 19.5 21 13 W43 L1 W29 L17 W24 W20 13 Martis, Tyler BRHS 12 1787 4 17 18 21 13 W42 L15 W24 L11 W18 W22 14 Plumb, Justin Rodney GCLACA 10 1700 4 16 17 20 13 W51 L32 W19 L20 W28 W27 15 Martinez, Isaac GLPREP 10 2159 3.5 21.5 24.5 27.5 16 W28 W13 D16 W4 L3 L5 16 Chen, Derek H CFHS 10 1965 3.5 21 23.5 26 15.5 W35 W7 D15 D2 D17 L6 17 Woodson, Tyler GILBHS 1640 3.5 19 19 17.5 14 W50 L5 W30 W12 D16 L7 18 Cancio, Aiya CFHS 9 1469 3.5 18.5 20 17.5 12.5 W36 D6 L4 W46 L13 W25 AZ Girls' Invitational Representative 19 Folden, Kurt CHANPR 10 1207 3 14 18 12 10 L32 W50 L14 W31 W23 L11 20 Thornton, -

Contemporary German Literature Collection) Brian Vetruba Washington University in St Louis, [email protected]
Washington University in St. Louis Washington University Open Scholarship University Libraries Publications University Libraries 2015 Twenty-ninth Annual Bibliography 2015 (Contemporary German Literature Collection) Brian Vetruba Washington University in St Louis, [email protected] Paul Michael Lützeler Washington University in St. Louis, [email protected] Katharina Böhm Washington University in St. Louis, [email protected] Follow this and additional works at: https://openscholarship.wustl.edu/lib_papers Part of the German Literature Commons, and the Library and Information Science Commons Recommended Citation Vetruba, Brian; Lützeler, Paul Michael; and Böhm, Katharina, "Twenty-ninth Annual Bibliography 2015 (Contemporary German Literature Collection)" (2015). University Libraries Publications. 20. https://openscholarship.wustl.edu/lib_papers/20 This Bibliography is brought to you for free and open access by the University Libraries at Washington University Open Scholarship. It has been accepted for inclusion in University Libraries Publications by an authorized administrator of Washington University Open Scholarship. For more information, please contact [email protected]. Max Kade Center for Contemporary German Literature Max Kade Zentrum für deutschsprachige Gegenwartsliteratur Director: Paul Michael Lützeler Twenty-ninth Annual Bibliography 2015 Editor: Brian W. Vetruba Editorial Assistant: Katharina Böhm February 28, 2017 Washington University in St. Louis Department of Germanic Languages and Literatures Max Kade Center -

Draft Project List 2017-04-24
Transportation System Development Charge (TSDC) Project List Total Non- Growth City SDC Eligible Project # Project Name Project Location Estimated Growth Cost Responsibility Cost Cost Cost Share Share Driving Solutions (Intersections, Extensions & Expansions) Molalla Avenue from Washington Street to Molalla Avenue/ Beavercreek Road Adaptive D1 Gaffney Lane; Beavercreek Road from Molalla $1,565,000 75% 25% 100% $391,250 Signal Timing Avenue to Maple Lane Road D2 Beavercreek Road Traffic Surveillance Molalla Avenue to Maple Lane Road $605,000 75% 25% 100% $151,250 D3 Washington Street Traffic Surveillance 7th Street to OR 213 $480,000 75% 25% 100% $120,000 D4 7th Street/Molalla Avenue Traffic Surveillance Washington Street to OR 213 $800,000 75% 25% 100% $200,000 OR 213/ 7th Street-Molalla Avenue/ D5 Washington Street Integrated Corridor I-205 to Henrici Road $1,760,000 75% 25% 30% $132,000 Management D6 OR 99E Integrated Corridor Management OR 224 (in Milwaukie) to 10th Street $720,000 75% 25% 30% $54,000 D7 14th Street Restriping OR 99E to John Adams Street $845,000 74% 26% 100% $216,536 D8 15th Street Restriping OR 99E to John Adams Street $960,000 80% 20% 100% $192,000 OR 213/Beavercreek Road Weather D9 OR 213/Beavercreek Road $120,000 100% 0% 30% $0 Information Station Warner Milne Road/Linn Avenue Road Weather D10 Warner Milne Road/Linn Avenue $120,000 100% 0% 100% $0 Information Station D11 Optimize existing traffic signals Citywide $50,000 75% 25% 100% $12,500 D12 Protected/permitted signal phasing Citywide $65,000 75% 25% 100% -

Likely to Be Funded Transportation System
Table 2: Likely to be Funded Transportation System Project # Project Description Project Extent Project Elements Priority Further Study Identify and evaluate circulation options to reduce motor OR 213/Beavercreek Road Refinement OR 213 from Redland Road to Molalla D0 vehicle congestion along the corridor. Explore alternative Short-term Plan Avenue mobility targets. Identify and evaluate circulation options to reduce motor I-205 at the OR 99E and OR 213 Ramp vehicle congestion at the interchanges. Explore alternative D00 I-205 Refinement Plan Short-term Terminals mobility targets, and consider impacts related to a potential MMA Designation for the Oregon City Regional Center. Driving Solutions (Intersection and Street Management- see Figure 16) Molalla Avenue from Washington Street to Molalla Avenue/ Beavercreek Road Deploy adaptive signal timing that adjusts signal timings to D1 Gaffney Lane; Beavercreek Road from Molalla Short-term Adaptive Signal Timing match real-time traffic conditions. Avenue to Maple Lane Road Option 1: Convert 14th Street to one-way eastbound between McLoughlin Boulevard and John Adams Street: • Convert the Main Street/14th Street intersection to all-way stop control (per project D13). • From McLoughlin Boulevard to Main Street, 14th Street would be restriped to include two 12-foot eastbound travel lanes, a six-foot eastbound bike lane, a six-foot westbound contra-flow bike lane, and an eight-foot landscaping buffer on the north side • From Main Street to Washington Street, 14th Street would be restriped to include -

FY 2010 Volume 1
DOE/CF-035 Volume 1 FY 2010 Congressional Budget Request National Nuclear Security Administration Office of the Administrator Weapons Activities Defense Nuclear Nonproliferation Naval Reactors Office of Chief Financial Officer May 2009 Volume 1 DOE/CF-035 Volume 1 FY 2010 Congressional Budget Request National Nuclear Security Administration Office of the Administrator Weapons Activities Defense Nuclear Nonproliferation Naval Reactors Office of Chief Financial Officer May 2009 Volume 1 Printed with soy ink on recycled paper Office of the Administrator Weapons Activities Defense Nuclear Nonproliferation Naval Reactors Office of the Administrator Weapons Activities Defense Nuclear Nonproliferation Naval Reactors Volume 1 Table of Contents Page Appropriation Account Summary.............................................................................................................3 NNSA Overview.......................................................................................................................................5 Office of the Administrator.....................................................................................................................25 Weapons Activities .................................................................................................................................47 Defense Nuclear Nonproliferation........................................................................................................345 Naval Reactors......................................................................................................................................461 -

NYISO Emergency Operations Manual
N Y I S O E M E R G E N C Y O P E R A T I O N S M A N U A L Table A.5 List of Thunderstorm Multiple Contingencies Cases Table A.5 List of Thunderstorm Multiple Contingencies Cases 1. F38, Y86, F39, Y87, Wood St. Bank 2, Pleasantville Bank 1, 311 2. F38, Y86, F39, Y87, Wood St. Bank 2, Pleasantville Bank 1, 77 3. F38, Y86, F39, Y87, Wood St. Bank 2, Pleasantville Bank 1, Y94, TA5, Bank (95891) 4. F38, Y86, F39, Y87, Wood St. Bank 2, Pleasantville Bank 1, Y88 5. F38, Y86, F39, Y87, Wood St. Bank 2, Pleasantville Bank 1, F31, W81 6. F38, Y86, F39, Y87, Wood St. Bank 2, Pleasantville Bank 1, W82, Eastview Bank 2S, W65 7. F38, Y86, F39, Y87, Wood St. Bank 2, Pleasantville Bank 1, W93, Eastview Bank 2N, W79 8. F38, Y86, F39, Y87, Wood St. Bank 2, Pleasantville Bank 1, A2253 9. F38, Y86, F39, Y87, Wood St. Bank 2, Pleasantville Bank 1, W75 10. F38, Y86, F39, Y87, Wood St. Bank 2, Pleasantville Bank 1, 301 11. F38, Y86, F39, Y87, Wood St. Bank 2, Pleasantville Bank 1, 303 12. W89, W73, W90, W74, Y50, Pleasantville Bank 2, 311 13. W89, W73, W90, W74, Y50, Pleasantville Bank 2, 77 14. W89, W73, W90, W74, Y50, Pleasantville Bank 2, Y94, TA5 Bank (95891) 15. W89, W73, W90, W74, Y50, Pleasantville Bank 2, Y88 16. W89, W73, W90, W74, Y50, Pleasantville Bank 2, F31, W81 17. W89, W73, W90, W74, Y50, Pleasantville Bank 2, W82 Eastview Bank 2S, W65 18. -

Nuclear Matters. a Practical Guide 5B
NUCLEAR MATTERS A Practical Guide Form Approved Report Documentation Page OMB No. 0704-0188 Public reporting burden for the collection of information is estimated to average 1 hour per response, including the time for reviewing instructions, searching existing data sources, gathering and maintaining the data needed, and completing and reviewing the collection of information. Send comments regarding this burden estimate or any other aspect of this collection of information, including suggestions for reducing this burden, to Washington Headquarters Services, Directorate for Information Operations and Reports, 1215 Jefferson Davis Highway, Suite 1204, Arlington VA 22202-4302. Respondents should be aware that notwithstanding any other provision of law, no person shall be subject to a penalty for failing to comply with a collection of information if it does not display a currently valid OMB control number. 1. REPORT DATE 3. DATES COVERED 2. REPORT TYPE 2008 00-00-2008 to 00-00-2008 4. TITLE AND SUBTITLE 5a. CONTRACT NUMBER Nuclear Matters. A Practical Guide 5b. GRANT NUMBER 5c. PROGRAM ELEMENT NUMBER 6. AUTHOR(S) 5d. PROJECT NUMBER 5e. TASK NUMBER 5f. WORK UNIT NUMBER 7. PERFORMING ORGANIZATION NAME(S) AND ADDRESS(ES) 8. PERFORMING ORGANIZATION Office of the Deputy Assistant to the Secretary of Defense (Nuclear REPORT NUMBER Matters),The Pentagon Room 3B884,Washington,DC,20301-3050 9. SPONSORING/MONITORING AGENCY NAME(S) AND ADDRESS(ES) 10. SPONSOR/MONITOR’S ACRONYM(S) 11. SPONSOR/MONITOR’S REPORT NUMBER(S) 12. DISTRIBUTION/AVAILABILITY STATEMENT Approved for public release; distribution unlimited 13. SUPPLEMENTARY NOTES 14. ABSTRACT 15. SUBJECT TERMS 16. SECURITY CLASSIFICATION OF: 17. -

Method for Scalar Multiplication
Sensors 2013, 13, 9483-9512; doi:10.3390/s130709483 OPEN ACCESS sensors ISSN 1424-8220 www.mdpi.com/journal/sensors Article Fixed-Base Comb with Window-Non-Adjacent Form (NAF) Method for Scalar Multiplication Hwajeong Seo 1, Hyunjin Kim 1, Taehwan Park 1, Yeoncheol Lee 1, Zhe Liu 2 and Howon Kim 1;* 1 Computer engineering, Pusan National University, Pusan 609-735, Korea; E-Mails: [email protected] (H.S.); [email protected] (H.K.); [email protected] (T.P.); [email protected] (Y.L.) 2 Laboratory of Algorithmics, Cryptology and Security, University of Luxembourg, 6, Rue Richard Coudenhove-Kalergi, Luxembourg L–1359, Luxembourg; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +82-51-510-1010; Fax: +82-51-517-2431. Received: 23 May 2013; in revised form: 12 July 2013 / Accepted: 16 July 2013 / Published: 23 July 2013 Abstract: Elliptic curve cryptography (ECC) is one of the most promising public-key techniques in terms of short key size and various crypto protocols. For this reason, many studies on the implementation of ECC on resource-constrained devices within a practical execution time have been conducted. To this end, we must focus on scalar multiplication, which is the most expensive operation in ECC. A number of studies have proposed pre-computation and advanced scalar multiplication using a non-adjacent form (NAF) representation, and more sophisticated approaches have employed a width-w NAF representation and a modified pre-computation table. In this paper, we propose a new pre-computation method in which zero occurrences are much more frequent than in previous methods. -

Product News
PRODUCT NEWS ROSS CONTROLS® Improves Low Temperature Ratings of Valves ¾ Expands Application Range ¾ Broadens Base of Potential Customers ¾ Compliments Circuits using Low Temp Series 21 Products ROSS CONTROLS® World Headquarters (November 2018) - Customers worldwide have long considered ROSS® products as best-in-class for extreme and harsh environments. ROSS® has earned a reputation for rugged reliability and long service life in these challenging applications. For decades ROSS® Series 21 valves have successfully operated in -40°F (-40°C) conditions. Similarly, many ROSS® legacy products have been effectively installed in circuits with the Series 21, even though the applications were well below the conservative temperature ratings published in our catalogs. It is well-known, water vapor in an air system has the potential to cause the formation of ice when operating at temperatures below +40°F (4°C). ROSS® has historically used this principle to guide the low temperature ratings of many legacy products instead of using the actual temperature capability of the valve design. Today, ROSS CONTROLS® is pleased to announce modernized low temperature ratings for many of our legacy products. Literature on the ROSS® website has been updated to reflect the lower temperature range for the valves on Page 2 of this announcement. No design or component changes have been made, so any valves on your shelf will operate at these lower temperatures. OEM business. Even if you sell ROSS® products in a warm climate where USER customers do not operate at low temperatures, you may have OEM customers who design & build equipment for cold weather applications outside of your territory. -

PC Cake Perform Pump
PC Cake Perform Pump Progressing cavity process pump, designed to be maintained in place without disconnecting from pipework. For pumping of highly viscous materials such as sludges, slurries, thick non-flowing pastes and dewatered sludge cake, in municipal and industrial process applications. Construction Materials of construction, available in cast iron, with a choice of rotor and stator materials to suit individual applications e.g. hard chrome plated rotor or natural rubber stator. Applications Typical applications for the PC cake perform pump include: • Heavy sludge cake transfer for greater than 30% dry solids concentration. • Dewatered and thickened sludge transfer. • Sludge blending. • Imported and organic waste sludge transfer. • Industrial process sludge with high percentage dry solids concentration. Performance Features Capacity, for flows up to 216 gpm (49 m3/h) and differential pressure • Maintain-in-place design allows for quick and easy removal of up to 350 psi (24 bar), to operate in a range of process temperatures rotating parts, and clearing of rag build-up, without disconnecting from 14 °F (-10 °C), up to 212 °F (100 °C). from pipework. • An auger screw conveyor for efficient feeding of the pump when Performance data handling high percentage dry solid sludge concentrations. • Gentle pumping action, minimises shear and crush damage to the 216 pumped product. GPM WA2 • Supplied with a baseplate to ease installation, or optional without. 176 • Fully sealed drive train to maximise life and minimise downtime. • Hard faced, single mechanical seal as standard, with packed W92 gland as an option. 150 • Designed to accommodate optional hopper or bridge breaker W82 W84 attachments.