SI Text Western Blot Analysis, Immunofluorescence, and Immunohistochem- Quantitative Real-Time PCR
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Learning from Cadherin Structures and Sequences: Affinity Determinants and Protein Architecture
Learning from cadherin structures and sequences: affinity determinants and protein architecture Klára Fels ıvályi Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2014 Klara Felsovalyi All rights reserved ABSTRACT Learning from cadherin structures and sequences: affinity determinants and protein architecture Klara Felsovalyi Cadherins are a family of cell-surface proteins mediating adhesion that are important in development and maintenance of tissues. The family is defined by the repeating cadherin domain (EC) in their extracellular region, but they are diverse in terms of protein size, architecture and cellular function. The best-understood subfamily is the type I classical cadherins, which are found in vertebrates and have five EC domains. Among the five different type I classical cadherins, the binding interactions are highly specific in their homo- and heterophilic binding affinities, though their sequences are very similar. As previously shown, E- and N-cadherins, two prototypic members of the subfamily, differ in their homophilic K D by about an order of magnitude, while their heterophilic affinity is intermediate. To examine the source of the binding affinity differences among type I cadherins, we used crystal structures, analytical ultracentrifugation (AUC), surface plasmon resonance (SPR), and electron paramagnetic resonance (EPR) studies. Phylogenetic analysis and binding affinity behavior show that the type I cadherins can be further divided into two subgroups, with E- and N-cadherin representing each. In addition to the affinity differences in their wild-type binding through the strand-swapped interface, a second interface also shows an affinity difference between E- and N-cadherin. -

Somatic Mutational Landscapes of Adherens Junctions and Their
1 Somatic mutational landscapes of adherens junctions and their 2 functional consequences in cutaneous melanoma development 3 4 Praveen Kumar Korla,1 Chih-Chieh Chen,2 Daniel Esguerra Gracilla,1 Ming-Tsung Lai,3 Chih- 5 Mei Chen,4 Huan Yuan Chen,5 Tritium Hwang,1 Shih-Yin Chen,4,6,* Jim Jinn-Chyuan Sheu1,4, 6-9,* 6 1Institute of Biomedical Sciences, National Sun Yat-sen University, Kaohsiung 80242, Taiwan; 7 2Institute of Medical Science and Technology, National Sun Yat-sen University, Kaohsiung 80424, 8 Taiwan; 3Department of Pathology, Taichung Hospital, Ministry of Health and Welfare, Taichung 9 40343, Taiwan; 4Genetics Center, China Medical University Hospital, Taichung 40447, Taiwan; 10 5Institute of Biomedical Sciences, Academia Sinica, Taipei 11574, Taiwan; 6School of Chinese 11 Medicine, China Medical University, Taichung 40402, Taiwan; 7Department of Health and 12 Nutrition Biotechnology, Asia University, Taichung 41354, Taiwan; 8Department of 13 Biotechnology, Kaohsiung Medical University, Kaohsiung 80708, Taiwan; 9Institute of 14 Biopharmaceutical Sciences, National Sun Yat-sen University, Kaohsiung 80242, Taiwan 15 16 PKK, CCC and DEG contributed equally to this study. 17 *Correspondence to: Dr. Shih-Yin Chen ([email protected]) at Genetics Center, China 18 Medical University Hospital, Taichung, 40447, TAIWAN; or Dr. Jim Jinn-Chyuan Sheu 19 ([email protected]) at Institute of Biomedical Sciences, National Sun Yat-sen 20 University, Kaohsiung City 80424, TAIWAN. 21 22 Running title: mutational landscape of cadherins in melanoma 1 23 Abstract 24 Cell-cell interaction in skin homeostasis is tightly controlled by adherens junctions (AJs). 25 Alterations in such regulation lead to melanoma development. -

Sterculic Acid Alters Adhesion Molecules Expression and Extracellular Matrix Compounds to Regulate Migration of Lung Cancer Cells
cancers Article Sterculic Acid Alters Adhesion Molecules Expression and Extracellular Matrix Compounds to Regulate Migration of Lung Cancer Cells Rafael Peláez * , Rodrigo Ochoa, Ana Pariente, Ángela Villanueva-Martínez, Álvaro Pérez-Sala and Ignacio M. Larráyoz * Biomarkers and Molecular Signaling Group, Neurodegeneration Area, Center for Biomedical Research of La Rioja (CIBIR), Piqueras 98, 26006 Logroño, Spain; [email protected] (R.O.); [email protected] (A.P.); [email protected] (Á.V.-M.); [email protected] (Á.P.-S.) * Correspondence: [email protected] (R.P.); [email protected] (I.M.L.); Tel.: +34-941-278-770 ((ext. 84866) (R.P.) & (ext. 89878) (I.M.L.)) Simple Summary: Sterculic acid (SA) is a naturally occurring lipid with SCD1 inhibitory activity, but it also modifies many other pathways and underlying gene expression. SCD upregulation has been associated with tumor aggressiveness and progression. Effects of SA treatment over extracellular matrix compounds and adhesion molecule expression have not been described in cancer cells up to now. Our results show that SA induces cell death at high dose, but we also observed that lower concentrations of SA treatments also reduce cell adhesion-migration and modify integrins and extracellular matrix compounds expression. Citation: Peláez, R.; Ochoa, R.; Abstract: Sterculic acid (SA) is a cyclopropenoid fatty acid isolated from Sterculia foetida seeds. Pariente, A.; Villanueva-Martínez, Á.; This molecule is a well-known inhibitor of SCD1 enzyme, also known as D9-desaturase, which Pérez-Sala, Á.; Larráyoz, I.M. main function is related to lipid metabolism. However, recent studies have demonstrated that Sterculic Acid Alters Adhesion it also modifies many other pathways and the underlying gene expression. -

Academia Arena 2019;11(1)
Academia Arena 2019;11(1) http://www.sciencepub.net/academia Cadherin Mark Herbert, PhD World Development Institute 39-06 Main Street, Flushing, Queens, New York 11354, USA, [email protected] Abstract: Cadherins (calcium dependent adhesion molecules) are a class of type-1 transmembrane proteins. They play important roles in cell adhesion, ensuring that cells within tissues are bound together. Their functions are dependent from calcium ions (Ca2+). The cadherin superfamily includes cadherins, protocadherins, desmogleins, and desmocollins, etc. In structure, they share cadherin repeats, which are the extracellular Ca2+-binding domains. There are multiple classes of cadherin molecule, each designated with a prefix (generally noting the type of tissue with which it is associated). It has been observed that cells containing a specific cadherin subtype tend to cluster together to the exclusion of other types, both in cell culture and during development. For example, cells containing N- cadherin tend to cluster with other N-cadherin expressing cells. However, it has been noted that the mixing speed in the cell culture experiments can have an effect on the extent of homotypic specificity. In addition, several groups have observed heterotypic binding affinity (i.e., binding of different types of cadherin together) in various assays. One current model proposes that cells distinguish cadherin subtypes based on kinetic specificity rather than thermodynamic specificity, as different types of cadherin homotypic bonds have different lifetimes. [Mark Herbert, PhD. Cadherin. Academ Arena 2019;11(1):166-168]. ISSN 1553-992X (print); ISSN 2158-771X (online). http://www.sciencepub.net/academia. 4. doi:10.7537/marsaaj110119.04. Keywords: cadherin; cell; protein; DNA; life Cadherins (calcium dependent adhesion have no cytoplasmic domains and must be tethered to molecules) are a class of type-1 transmembrane the plasma membrane. -
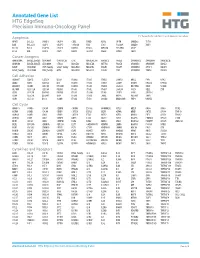
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel For Research Use Only. Not for use in diagnostic procedures. Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 -

Modulation of Skeletal Muscle Insulin Signaling with Chronic Caloric Restriction in Cynomolgus Monkeys
Diabetes Publish Ahead of Print, published online March 31, 2009 Modulation of skeletal muscle insulin signaling with chronic caloric restriction in cynomolgus monkeys Zhong Q. Wang1 Elizabeth Floyd1 Jianhua Qin1 Xiaotuan Liu1 Yongmei Yu1 Xian H Zhang1 Janice D. Wagner2 William T. Cefalu1 Division of Nutrition and Chronic Diseases1, Pennington Biomedical Research Center, Louisiana State University System; Department of Pathology, Wake Forest University School of Medicine 2 Corresponding Author: William T Cefalu, M.D. Email: [email protected] Submitted 18 July 2008 and accepted 19 March 2009. This is an uncopyedited electronic version of an article accepted for publication in Diabetes. The American Diabetes Association, publisher of Diabetes, is not responsible for any errors or omissions in this version of the manuscript or any version derived from it by third parties. The definitive publisher-authenticated version will be available in a future issue of Diabetes in print and online at http://diabetes.diabetesjournals.org. 1 Copyright American Diabetes Association, Inc., 2009 Objective: Caloric restriction (CR) has been shown to retard aging processes, extend maximal life span, and consistently increase insulin action in experimental animals. The mechanism by which CR enhances insulin action, specifically in higher species, is not precisely known. We sought to examine insulin receptor signaling and transcriptional alterations in skeletal muscle of nonhuman primates subjected to caloric restriction over a 4 year period. Research Design: After baseline, 32 male adult cynomolgus monkeys (Macaca fascicularis) were randomized to an Ad libitum diet (AL) or to 30% CR. Dietary intake, body weight and insulin sensitivity were obtained at routine intervals over 4 years. -

Pan-Cancer Genomic Amplifications Underlie a Wnt Hyperactivation Phenotype
bioRxiv preprint doi: https://doi.org/10.1101/519611; this version posted January 13, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Pan-cancer genomic amplifications underlie a Wnt hyperactivation phenotype 2 associated with stem cell-like features leading to poor prognosis 3 4 5 6 Wai Hoong Chang and Alvina G. Lai 7 8 9 Nuffield Department of Medicine, University of Oxford, 10 Old Road Campus, Oxford, OX3 7FZ, United Kingdom 11 12 For correspondence: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/519611; this version posted January 13, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 13 List of Abbreviations 14 TCGA The Cancer Genome Atlas KEGG Kyoto Encyclopedia of Genes and Genomes GO Gene Ontology ROC Receiver operating characteristic AUC Area under the curve HR Hazard ratio TNM Tumor, node and metastasis HIF Hypoxia inducible factor TF Transcription factor EMT Epithelial-to-mesenchymal transition 15 bioRxiv preprint doi: https://doi.org/10.1101/519611; this version posted January 13, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Elevated Frequency of Loss of Heterozygosity in Mammary Tumors Arising in Mouse Mammary Tumor Virus/Neu Transgenic Mice1
[CANCER RESEARCH 59, 2438–2444, May 15, 1999] Elevated Frequency of Loss of Heterozygosity in Mammary Tumors Arising in Mouse Mammary Tumor Virus/neu Transgenic Mice1 Marc Cool and Paul Jolicoeur2 Laboratory of Molecular Biology, Clinical Research Institute of Montreal, Montreal, Quebec, H2W 1R7 Canada [M. C., P. J.]; Department of Microbiology and Immunology, University of Montreal, Montreal, Quebec, H3C 3J7 Canada [P. J.]; and Division of Experimental Medicine, McGill University, Montreal, Quebec, H3G 1A4 Canada [P. J.] ABSTRACT LOH. The number and identity of most tumor suppressor genes involved in human breast cancers are unknown, and only a few of Loss of heterozygosity (LOH) analysis was performed on 62 mammary them have been identified; p53 (16), RB1 (17, 18), PTEN/MMAC1 tumors that were induced in (BALB/c 3 C57BL/6)F mouse mammary 1 (19, 20), E-cadherin (21, 22), FHIT (23, 24), TSG101 (25), and tumor virus/neu transgenic mice. Eighty-six simple sequence length poly- INK4a morphism markers were used to cover all of the somatic chromosomes. p16 (26, 27) have all been found to be inactivated at various Frequency of LOH was observed to be significant for chromosomes 4 frequencies in sporadic breast carcinoma. In familial breast cancer, (50%), 19 (32%), and 8 (21%). On chromosome 4, at least three distinct inactivation of BRCA1 and BRCA2 is a relatively frequent event regions of allelic deletions could be identified: one proximal to 22 cM; the (28, 29). second close to the p16INK4a/p15INK4b locus, which is commonly deleted in To study the molecular nature of the erbB-2/neu transformation various tumors; and the third one in the proximity of Mom1. -

CDH15 Monoclonal Antibody, Clone 21G4
CDH15 monoclonal antibody, clone Storage Instruction: Store at 4°C on dry atmosphere. 21G4 After reconstitution with deionized water, store at -20°C or lower. Catalog Number: MAB3922 Aliquot to avoid repeated freezing and thawing. Regulatory Status: For research use only (RUO) Entrez GeneID: 1013 Product Description: Mouse monoclonal antibody Gene Symbol: CDH15 raised against partial recombinant CDH15. Gene Alias: CDH14, CDH3, CDHM, MCAD Clone Name: 21G4 Gene Summary: This gene is a member of the cadherin Immunogen: Recombinant protein corresponding to superfamily of genes, encoding calcium-dependent extracellular region of human CDH15. intercellular adhesion glycoproteins. Cadherins consist of an extracellular domain containing 5 cadherin Host: Mouse domains, a transmembrane region, and a conserved cytoplasmic domain. Transcripts from this particular Reactivity: Human cadherin are expressed in myoblasts and upregulated in myotubule-forming cells. The protein is thought to be Applications: EIA, IF, WB-Tr essential for the control of morphogenetic processes, (See our web site product page for detailed applications specifically myogenesis, and may provide a trigger for information) terminal muscle cell differentiation. [provided by RefSeq] Protocols: See our web site at http://www.abnova.com/support/protocols.asp or product page for detailed protocols Specificity: Cadherins are a family of transmembrane glycoproteins that play a key role in Calcium dependent cell-cell adhesion. Several members of the cadherin family have been identified so far, including E- (epithelial), P- (placental), N- (neuronal) and M- (muscle) cadherin. M-Cadherin is expressed in myoblasts and might be involved in skeletal muscle cell differentiation. Does not cross-react with mouse. Form: Lyophilized Isotype: IgG1 Recommend Usage: ELISA (0.05 ug/mL) Western Blot (1 ug/mL for AP/BCIP/NBT (MTT) detection 0.5 ug/mL for HRP/ECL detection) Immunofluorescence (5 ug/mL) The optimal working dilution should be determined by the end user. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN.