CHRO Guidebook
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Food Or Beverage Product, Or Probiotic Composition, Comprising Lactobacillus Johnsonii 456
(19) TZZ¥¥¥ _T (11) EP 3 536 328 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 11.09.2019 Bulletin 2019/37 A61K 35/74 (2015.01) A61K 35/66 (2015.01) A61P 35/00 (2006.01) (21) Application number: 19165418.5 (22) Date of filing: 19.02.2014 (84) Designated Contracting States: • SCHIESTL, Robert, H. AL AT BE BG CH CY CZ DE DK EE ES FI FR GB Encino, CA California 91436 (US) GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO • RELIENE, Ramune PL PT RO RS SE SI SK SM TR Los Angeles, CA California 90024 (US) • BORNEMAN, James (30) Priority: 22.02.2013 US 201361956186 P Riverside, CA California 92506 (US) 26.11.2013 US 201361909242 P • PRESLEY, Laura, L. Santa Maria, CA California 93458 (US) (62) Document number(s) of the earlier application(s) in • BRAUN, Jonathan accordance with Art. 76 EPC: Tarzana, CA California 91356 (US) 14753847.4 / 2 958 575 (74) Representative: Müller-Boré & Partner (71) Applicant: The Regents of the University of Patentanwälte PartG mbB California Friedenheimer Brücke 21 Oakland, CA 94607 (US) 80639 München (DE) (72) Inventors: Remarks: • YAMAMOTO, Mitsuko, L. This application was filed on 27-03-2019 as a Alameda, CA California 94502 (US) divisional application to the application mentioned under INID code 62. (54) FOOD OR BEVERAGE PRODUCT, OR PROBIOTIC COMPOSITION, COMPRISING LACTOBACILLUS JOHNSONII 456 (57) The present invention relates to food products, beverage products and probiotic compositions comprising Lacto- bacillus johnsonii 456. EP 3 536 328 A1 Printed by Jouve, 75001 PARIS (FR) EP 3 536 328 A1 Description CROSS-REFERENCE TO RELATED APPLICATIONS 5 [0001] This application claims the benefit of U.S. -

Genomics of Helicobacter Species 91
Genomics of Helicobacter Species 91 6 Genomics of Helicobacter Species Zhongming Ge and David B. Schauer Summary Helicobacter pylori was the first bacterial species to have the genome of two independent strains completely sequenced. Infection with this pathogen, which may be the most frequent bacterial infec- tion of humanity, causes peptic ulcer disease and gastric cancer. Other Helicobacter species are emerging as causes of infection, inflammation, and cancer in the intestine, liver, and biliary tract, although the true prevalence of these enterohepatic Helicobacter species in humans is not yet known. The murine pathogen Helicobacter hepaticus was the first enterohepatic Helicobacter species to have its genome completely sequenced. Here, we consider functional genomics of the genus Helico- bacter, the comparative genomics of the genus Helicobacter, and the related genera Campylobacter and Wolinella. Key Words: Cytotoxin-associated gene; H-Proteobacteria; gastric cancer; genomic evolution; genomic island; hepatobiliary; peptic ulcer disease; type IV secretion system. 1. Introduction The genus Helicobacter belongs to the family Helicobacteriaceae, order Campylo- bacterales, and class H-Proteobacteria, which is also known as the H subdivision of the phylum Proteobacteria. The H-Proteobacteria comprise of a relatively small and recently recognized line of descent within this extremely large and phenotypically diverse phy- lum. Other genera that colonize and/or infect humans and animals include Campylobac- ter, Arcobacter, and Wolinella. These organisms are all microaerophilic, chemoorgano- trophic, nonsaccharolytic, spiral shaped or curved, and motile with a corkscrew-like motion by means of polar flagella. Increasingly, free living H-Proteobacteria are being recognized in a wide range of environmental niches, including seawater, marine sedi- ments, deep-sea hydrothermal vents, and even as symbionts of shrimp and tubeworms in these environments. -

Co-Infection Associated with Diarrhea in a Colony of <I>Scid
Laboratory Animal Science Vol 48, No 5 Copyright 1998 October 1998 by the American Association for Laboratory Animal Science Helicobacter bilis/Helicobacter rodentium Co-Infection Associated with Diarrhea in a Colony of scid Mice Nirah H. Shomer,* Charles A. Dangler, Robert P. Marini, and James G. Fox† Abstract _ An outbreak of diarrhea spanning 3 months occurred in a breeding colony of scid/Trp53 knockout mice. Approximately a third of the 150 mice were clinically affected, with signs ranging from mucoid or watery diarrhea to severe hemorrhagic diarrhea with mortality. Helicobacter bilis and the newly recognized urease-negative organ- ism H. rodentium were isolated from microaerobic culture of feces or cecal specimens from affected mice. Dual infection with H. bilis and H. rodentium were confirmed by culture and polymerase chain reaction (PCR) in several animals. Both Helicobacter species rapidly colonized immunocompetent sentinel mice exposed to bedding from cages containing affected mice, but the sentinel remained asymptomatic. Mice with diarrhea had multifocal to segmental proliferative typhlitis, colitis, and proctitis. Several affected mice had multifocal mucosal necrosis with a few focal ulcers in the cecum, colon, and rectum. Mice with diarrhea were treated with antibiotic food wafers (1.5 mg of amoxicillin, 0.69 mg of metronidazole, and 0.185 mg of bismuth/mouse per day) previously shown to eradi- cate H. hepaticus in immunocompetent mice. Antibiotic treatment resulted in resolution of diarrhea, but not eradication of H. bilis and H. rodentium; mice continued to have positive PCR results after a 2-week treatment regimen, and clinical signs of diarrhea returned in some mice when treatment was suspended. -

Enterohepatic Lesions in SCID Mice Infected with Helicobacter Bilis
Laboratory Animal Science Vol 48, No 4 Copyright 1998 August 1998 by the American Association for Laboratory Animal Science Enterohepatic Lesions in SCID Mice Infected with Helicobacter bilis Craig L. Franklin, Lela K. Riley, Robert S. Livingston, Catherine S. Beckwith, Cynthia L. Besch-Williford, and Reuel R. Hook, Jr. Abstract _ Helicobacter bilis is a recently identified species that colonizes the intestine and liver of mice. In immunocompetent mice, infections have been associated with mild hepatitis, and in immunocompromised mice, inflammatory bowel disease has been induced by intraperitoneal inoculation of the organism. We re- port inoculation of 6-week-old C.B-17 scid/scid mice by gastric gavage with approximately 107 H. bilis colony- forming units. Groups of mice were euthanized and necropsied 12, 24, and 36 weeks after inoculation. Mild to moderate proliferative typhlitis was evident in all mice at 12 and 36 weeks after inoculation and in most mice 24 weeks after inoculation. Mild to severe chronic active hepatitis was detected in 10 of 10 male mice and 3 of 10 female mice. These results indicate that H. bilis can cause moderate to severe enterohepatic disease in immunocompromised mice. The genus Helicobacter is a rapidly expanding genus volved in lesion development. Culture of specimens from currently containing 17 named species. Members of this mice confirmed intestinal colonization with H. hepaticus. Fox genus are microaerophilic, have curved to spiral rod mor- et al. reported enteric lesions in immunocompetent germ- phology, and are motile by flagella that vary in number free Swiss Webster mice infected with H. hepaticus (15), and and location among various species (1). -

Biliary Tract Microbiota: a New Kid on the Block of Liver Diseases?
European Review for Medical and Pharmacological Sciences 2020; 24: 2750-2775 Biliary tract microbiota: a new kid on the block of liver diseases? A. NICOLETTI1, F.R. PONZIANI2, E. NARDELLA1, G. IANIRO2, A. GASBARRINI1, L. ZILERI DAL VERME2 1Internal Medicine, Gastroenterology and Hepatology, Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Università Cattolica del Sacro Cuore, Rome, Italy 2Internal Medicine, Gastroenterology and Hepatology, Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Rome, Italy Abstract. – The microbiome plays a crucial man body1,2. Indeed, a resident microbiota has recent- role in maintaining the homeostasis of the or- ly been described in several human environments ganism. Recent evidence has provided novel previously described as devoid of microorganisms, insights for understanding the interaction be- such as the urinary tract and the stomach3-9. Even tween the microbiota and the host. However, the 10 vast majority of such studies have analyzed the healthy placenta hosts microbial communities . interactions taking place in the intestinal tract. Bile has traditionally been considered sterile The biliary tree has traditionally been consid- under normal conditions11-14. ered sterile under normal conditions. However, The physical and chemical features of bile and the advent of metagenomic techniques has re- its antimicrobial activity were supposed to create vealed an unexpectedly rich bacterial communi- a hostile environment for bacteria. Moreover, the ty in the biliary tract. Associations between specific microbiolog- difficulty in collecting bile samples, coupled with ical patterns and inflammatory biliary diseases the lack of sensibility of culture techniques in and cancer have been recently described. Hence, detecting microbes in low-charge samples, sus- biliary dysbiosis may be a primary trigger in the tained this hypothesis for a long time. -
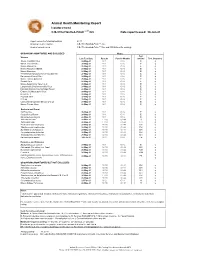
2021 ARC Health Report
Animal Health Monitoring Report Isolator reared C.B-17/IcrHanHsd-Prkdc scid /Arc Date report issued: 30-Jun-21 Report covers the following isolators: RI 27 Strains present in isolator: C.B-17/IcrHanHsd-Prkdc scid /Arc Strain of animal tested: C.B-17/IcrHanHsd-Prkdc scid /Arc and ICR Outbred for serology ORGANISMS MONITORED AND EXCLUDED Mouse Test Viruses Last Test Date Results Past 18 Months method Test frequency Mouse Hepatitis Virus 26-May-21 0 / 1 0 / 6 E q Minute Virus of Mice 26-May-21 0 / 1 0 / 6 E q Mouse Parvovirus 26-May-21 0 / 1 0 / 6 E q Murine Rotavirus (EDIM) 26-May-21 0 / 1 0 / 6 E q Mouse Norovirus 26-May-21 0 / 1 0 / 6 E q Theiler's Encephalomyelitis Virus (GD VII) 26-May-21 0 / 1 0 / 6 E q Pneumonia Virus of Mice 26-May-21 0 / 1 0 / 6 E q Murine Cytomegalovirus 26-May-21 0 / 1 0 / 6 E q Sendai Virus 26-May-21 0 / 1 0 / 6 E q Mouse Adenovirus Type 1 & 2 26-May-21 0 / 1 0 / 6 E q Lymphocytic Choriomeningitis Virus 26-May-21 0 / 1 0 / 6 E q Hantaan (Korean Haemorrhagic Fever) 26-May-21 0 / 1 0 / 6 E q Ectromelia (Mousepox) Virus 26-May-21 0 / 1 0 / 6 E q Reovirus -3 26-May-21 0 / 1 0 / 6 E q Polyoma Virus 26-May-21 0 / 1 0 / 6 E q K Virus 26-May-21 0 / 1 0 / 6 E q Lactic Dehydrogenase Elevating Virus 26-May-21 0 / 1 0 / 6 E q Mouse Thymic Virus 26-May-21 0 / 1 0 / 6 E q 00-Jan-00 Bacteria and Fungi CAR bacillus 26-May-21 0 / 1 0 / 6 E q Clostridium piliforme 26-May-21 0 / 1 0 / 6 E q Mycoplasma pulmonis 26-May-21 0 / 1 0 / 6 E q Helicobacter spp.1 26-May-21 0 / 10 0 / 44 H q Salmonella spp. -
R Graphics Output
883 | Desulfovibrio vulgaris | DvMF_2825 298701 | Desulfovibrio | DA2_3337 1121434 | Halodesulfovibrio aestuarii | AULY01000007_gene1045 207559 | Desulfovibrio alaskensis | Dde_0991 935942 | Desulfonatronum lacustre | KI912608_gene2193 159290 | Desulfonatronum | JPIK01000018_gene1259 1121448 | Desulfovibrio gigas | DGI_0655 1121445 | Desulfovibrio desulfuricans | ATUZ01000018_gene2316 525146 | Desulfovibrio desulfuricans | Ddes_0159 665942 | Desulfovibrio | HMPREF1022_02168 457398 | Desulfovibrio | HMPREF0326_00453 363253 | Lawsonia intracellularis | LI0397 882 | Desulfovibrio vulgaris | DVU_0784 1121413 | Desulfonatronovibrio hydrogenovorans | JMKT01000008_gene1463 555779 | Desulfonatronospira thiodismutans | Dthio_PD0935 690850 | Desulfovibrio africanus | Desaf_1578 643562 | Pseudodesulfovibrio aespoeensis | Daes_3115 1322246 | Pseudodesulfovibrio piezophilus | BN4_12523 641491 | Desulfovibrio desulfuricans | DND132_2573 1121440 | Desulfovibrio aminophilus | AUMA01000002_gene2198 1121456 | Desulfovibrio longus | ATVA01000018_gene290 526222 | Desulfovibrio salexigens | Desal_3460 1121451 | Desulfovibrio hydrothermalis | DESAM_21057 1121447 | Desulfovibrio frigidus | JONL01000008_gene3531 1121441 | Desulfovibrio bastinii | AUCX01000006_gene918 1121439 | Desulfovibrio alkalitolerans | dsat_0220 941449 | Desulfovibrio | dsx2_0067 1307759 | Desulfovibrio | JOMJ01000003_gene2163 1121406 | Desulfocurvus vexinensis | JAEX01000012_gene687 1304872 | Desulfovibrio magneticus | JAGC01000003_gene2904 573370 | Desulfovibrio magneticus | DMR_04750 -

Helicobacter Pylori in Thai Patients with Cholangiocarcinoma and Its Association with Biliary Inflammation and Proliferation
DOI:10.1111/j.1477-2574.2011.00423.x HPB ORIGINAL ARTICLE Helicobacter pylori in Thai patients with cholangiocarcinoma and its association with biliary inflammation and proliferation Wongwarut Boonyanugomol1,5, Chariya Chomvarin1,5, Banchob Sripa2,5, Vajarabhongsa Bhudhisawasdi3,5, Narong Khuntikeo3,5, Chariya Hahnvajanawong1,5 & Amporn Chamsuwan4 1Department of Microbiology, 2Department of Pathology, 3Department of Surgery, 4Department of Forensic Medicine and 5Liver Fluke and Cholangiocarcinoma Research Center, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand Abstracthpb_423 177..184 Objectives: To investigate whether Helicobacter spp. infection and the cagA of H. pylori are associated with hepatobiliary pathology, specifically biliary inflammation, cell proliferation and cholangiocarcinoma (CCA). Methods: Helicobacter species including H. pylori, H. bilis and H. hepaticus were detected in the speci- mens using the polymerase chain reaction (PCR). Biliary inflammation of the liver and gallbladders was semi-quantitatively graded on hematoxylin and eosin (H&E)-stained slides. Biliary proliferation was evaluated by immunohistochemistry using the Ki-67-labelling index. Results: Helicobacter pylori was found in 66.7%, 41.5% and 25.0% of the patients in the CCA, cholelithiasis and control groups (P < 0.05), respectively. By comparison, H. bilis was found in 14.9% and 9.4% of the patients with CCA and cholelithiasis, respectively (P > 0.05), and was absent in the control group. The cagA gene of H. pylori was detected in 36.2% and 9.1% of the patients with CCA and cholelithiasis, respectively (P < 0.05). Among patients with CCA, cell inflammation and proliferation in the liver and gallbladder were significantly higher among those DNA H. -

Infection with Helicobacter Pylori Induces Epithelial to Mesenchymal Transition in Human Cholangiocytes
pathogens Article Infection with Helicobacter pylori Induces Epithelial to Mesenchymal Transition in Human Cholangiocytes Prissadee Thanaphongdecha 1,2, Shannon E. Karinshak 1, Wannaporn Ittiprasert 1, Victoria H. Mann 1, Yaovalux Chamgramol 3, Chawalit Pairojkul 3, James G. Fox 4, Sutas Suttiprapa 2, Banchob Sripa 2,3,* and Paul J. Brindley 1,* 1 Research Center for Neglected Tropical Diseases of Poverty, Department of Microbiology, Immunology and Tropical Medicine, School of Medicine & Health Sciences, The George Washington University, Washington, DC 20037, USA; [email protected] (P.T.); [email protected] (S.E.K.); [email protected] (W.I.); [email protected] (V.H.M.) 2 Tropical Disease Research Laboratory, Faculty of Medicine, Khon Kaen University, Khon Kaen 40002, Thailand; [email protected] 3 Department of Pathology, Faculty of Medicine, Khon Kaen University, Khon Kaen 40002, Thailand; [email protected] (Y.C.); [email protected] (C.P.) 4 Division of Comparative Medicine, Massachusetts Institute of Technology, Cambridge, MA 02139, USA; [email protected] * Correspondence: [email protected] (B.S.); [email protected] (P.J.B.) Received: 4 October 2020; Accepted: 18 November 2020; Published: 21 November 2020 Abstract: Recent reports suggest that the East Asian liver fluke infection, caused by Opisthorchis viverrini, which is implicated in opisthorchiasis-associated cholangiocarcinoma, serves as a reservoir of Helicobacter pylori. The opisthorchiasis-affected cholangiocytes that line the intrahepatic biliary tract are considered to be the cell of origin of this malignancy. Here, we investigated interactions in vitro among human cholangiocytes, Helicobacter pylori strain NCTC 11637, and the congeneric bacillus, Helicobacter bilis. Exposure to increasing numbers of H. -

Spiral Bacteria in the Human Stomach: the Gastric Helicobacters Andre Dubois, M.D., Ph.D
Synopses Spiral Bacteria in the Human Stomach: The Gastric Helicobacters Andre Dubois, M.D., Ph.D. Digestive Diseases Division, Department of Medicine, Uniformed Services University of the Health Sciences, Bethesda, Maryland, USA During the past decade, Helicobacter pylori has become recognized as one of the most common human pathogens, colonizing the gastric mucosa of almost all persons exposed to poor hygienic conditions from childhood. It also is often found, albeit with a lower frequency, in groups of high socioeconomic status. H. pylori causes chronic active gastritis and is a major factor in the pathogenesis of duodenal ulcers and, to a lesser extent, gastric ulcers. In addition, the presence of this bacterium is now recognized as a risk factor for gastric adenocarcinoma and lymphoma. Nevertheless, most infections appear without clinical consequences. In this second decade of intensive research, it is important to understand why H. pylori is sometimes a dangerous pathogen, and to determine how it can be eradicated in those at highest risk for severe disease. At the end of the 19th century, several types of Furthermore, in June 1994, the International spirochetes and spirilla were observed for the first Agency for Research on Cancer Working Group time in the stomach of animals (1,2). Beginning at stated , “H. pylori plays a causal role in the chain of the turn of the 20th century, similar spiral bacteria events leading to cancer,” referring to adenocarci- were found in gastrectomy specimens from patients noma and lymphoma of the stomach as well as to the with gastric cancer and peptic ulcer disease (3,4). -

Development of a Gut Microbiota Diagnostic Tool for Pediatric Inflammatory Bowel Disease Based on GA-Maptm Technology Platform
Development of a gut microbiota diagnostic tool for pediatric inflammatory bowel disease based on GA-mapTM technology platform DINA LILLESETH VANGEN Norwegian University of Life Science Department of Chemistry, Biotechnology and Food Science Master Thesis 2011/2012 ! ""! ! ABSTRACT Inflammatory bowel disease (IBD) is an idiopathic, severe disease, which is characterized by chronic inflammation of the gastrointestinal tract. The incidence of IBD has increased through the last decades and specially among the pediatric population. The time from onset of symptoms to a final diagnose is made, is often related to delays and for many patients it is an emotionally demanding process. Early investigation in suspected cases may reduce the delay so that a treatment can begin as soon as possible. The involvement of intestinal microflora for pathogenesis of IBD is a link to further investigations to understand the disease, and to help people who suffer from IBD. The aim of the present work was to distinguish between pediatric IBD and non-IBD by identifying signatures in the microbiota. This was accomplished by use of a diagnostic tool based on GA-mapTM technology and the use of single nucleotide primer extension (SNuPE) probes to search for complementary bacterial 16S rRNA gene sequences. Seventy-four feces samples were collected from cohort and tested against 77 SNuPE probes. Statistical analysis was performed with Partial Least Squares – Discriminant Analysis and presented specificity by 82 % and sensitivity by 86 %. Classification error presented 16 % and indicated how many that was misclassified by the model. Inflammatory bowel disease is considered to include two major disorders where Crohn’s disease is one of them, and best correlation was found between Crohn’s disease and non-IBD through statistical analysis. -
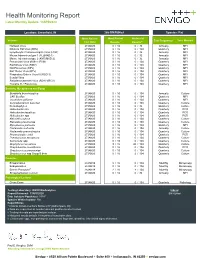
Health Monitoring Report Latest Monthly Update: 13APR2020
Health Monitoring Report Latest Monthly Update: 13APR2020 Location: Greenfield, IN 266-BN/RijHsd Species: Rat Most Recent Most Recent Historical Viruses Test Frequencyc Test Method Test Date Resultsa Resultsa,e Hantaan Virus 27JAN20 ## 0 / 16 0 / 32 Annually MFI Kilham's Rat Virus (KRV) 27JAN20 ## 0 / 16 0 / 104 Quarterly MFI Lymphocytic Choriomeningitis Virus (LCM) 27JAN20 ## 0 / 16 0 / 32 Annually MFI Mouse Adenovirus type 1 (FL)(MAD-1) 27JAN20 ## 0 / 16 0 / 32 Annually MFI Mouse Adenovirus type 2 (K87)(MAD-2) 27JAN20 ## 0 / 16 0 / 32 Annually MFI Pneumonia Virus of Mice (PVM) 27JAN20 ## 0 / 16 0 / 104 Quarterly MFI Rat Minute Virus (RMV) 27JAN20 ## 0 / 16 0 / 104 Quarterly MFI Rat Parvovirus (RPV) 27JAN20 ## 0 / 16 0 / 104 Quarterly MFI Rat Theiler Virus (RTV) 27JAN20 ## 0 / 16 0 / 104 Quarterly MFI Respiratory Enteric Virus III (REO 3) 27JAN20 ## 0 / 16 0 / 104 Quarterly MFI Sendai Virus 27JAN20 ## 0 / 16 0 / 104 Quarterly MFI Sialodacryoadenitis Virus (SDAV)(RCV) 27JAN20 ## 0 / 16 0 / 104 Quarterly MFI Toolan's H-1 Parvovirus 27JAN20 ## 0 / 16 0 / 104 Quarterly MFI Bacteria, Mycoplasma and Fungi Bordetella bronchiseptica 27JAN20 ## 0 / 16 0 / 104 Annually Culture CAR Bacillus 27JAN20 ## 0 / 16 0 / 104 Quarterly MFI Clostridium piliforme 27JAN20 ## 0 / 16 0 / 104 Quarterly MFI Corynebacterium kutscheri 27JAN20 ## 0 / 16 0 / 104 Quarterly Culture Dermatophytes 27JAN20 ## 0 / 16 0 / 32 Quarterly Culture Helicobacter bilis 27JAN20 ## 0 / 16 0 / 104 Quarterly PCR Helicobacter hepaticus 27JAN20 ## 0 / 16 0 / 104 Quarterly PCR Helicobacter