Biliary Tract Microbiota: a New Kid on the Block of Liver Diseases?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Umbilical Hernia with Cholelithiasis and Hiatal Hernia
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Springer - Publisher Connector Yamanaka et al. Surgical Case Reports (2015) 1:65 DOI 10.1186/s40792-015-0067-8 CASE REPORT Open Access Umbilical hernia with cholelithiasis and hiatal hernia: a clinical entity similar to Saint’striad Takahiro Yamanaka*, Tatsuya Miyazaki, Yuji Kumakura, Hiroaki Honjo, Keigo Hara, Takehiko Yokobori, Makoto Sakai, Makoto Sohda and Hiroyuki Kuwano Abstract We experienced two cases involving the simultaneous presence of cholelithiasis, hiatal hernia, and umbilical hernia. Both patients were female and overweight (body mass index of 25.0–29.9 kg/m2) and had a history of pregnancy and surgical treatment of cholelithiasis. Additionally, both patients had two of the three conditions of Saint’s triad. Based on analysis of the pathogenesis of these two cases, we consider that these four diseases (Saint’s triad and umbilical hernia) are associated with one another. Obesity is a common risk factor for both umbilical hernia and Saint’s triad. Female sex, older age, and a history of pregnancy are common risk factors for umbilical hernia and two of the three conditions of Saint’s triad. Thus, umbilical hernia may readily develop with Saint’s triad. Knowledge of this coincidence is important in the clinical setting. The concomitant occurrence of Saint’s triad and umbilical hernia may be another clinical “tetralogy.” Keywords: Saint’s triad; Cholelithiasis; Hiatal hernia; Umbilical hernia Background of our knowledge, no previous reports have described the Saint’s triad is characterized by the concomitant occur- coexistence of umbilical hernia with any of the three con- rence of cholelithiasis, hiatal hernia, and colonic diverticu- ditions of Saint’s triad. -

Cirrhosis of Liver Is a Risk Factor for Gallstone Disease
International Journal of Research in Medical Sciences Shirole NU et al. Int J Res Med Sci. 2017 May;5(5):2053-2056 www.msjonline.org pISSN 2320-6071 | eISSN 2320-6012 DOI: http://dx.doi.org/10.18203/2320-6012.ijrms20171841 Original Research Article Cirrhosis of liver is a risk factor for gallstone disease Nikhil U. Shirole*, Sudhir J. Gupta, Dharmesh K. Shah, Nitin R. Gaikwad, Tushar H. Sankalecha, Harit G. Kothari Department of Gastroenterology, Government Medical College and Super-speciality Hospital, Nagpur, Maharashtra, India Received: 07 February 2017 Revised: 22 March 2017 Accepted: 24 March 2017 *Correspondence: Dr. Nikhil U. Shirole, E-mail: [email protected] Copyright: © the author(s), publisher and licensee Medip Academy. This is an open-access article distributed under the terms of the Creative Commons Attribution Non-Commercial License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited. ABSTRACT Background: Gallstones are common clinical finding in general population. Mean prevalence rate in Indian population is 4-5%. The prevalence of gallstones is found to be high in cirrhotic patients compared to the general population in some western studies. Cause of this increased prevalence however is not known. Aim of the study was to evaluate prevalence of the gall stones in the cirrhotic patients, assess risk factors in cirrhotic patients and clinical presentation. Methods: This is the cross sectional observational study, included cirrhotic patients (compensated or decompensated). Risk factors for gallstone formation (age, gender and diabetes mellitus), characteristics of liver cirrhosis (etiology, Child Turcotte Pugh class, hypersplenism and varices) and clinical presentation were assessed in all cirrhotic patients with gallstones. -

Fecal Microbiota Transplant from Human to Mice Gives Insights Into the Role of the Gut Microbiota in Non-Alcoholic Fatty Liver Disease (NAFLD)
microorganisms Article Fecal Microbiota Transplant from Human to Mice Gives Insights into the Role of the Gut Microbiota in Non-Alcoholic Fatty Liver Disease (NAFLD) Sebastian D. Burz 1,2 , Magali Monnoye 1, Catherine Philippe 1, William Farin 3 , Vlad Ratziu 4, Francesco Strozzi 3, Jean-Michel Paillarse 3, Laurent Chêne 3, Hervé M. Blottière 1,2 and Philippe Gérard 1,* 1 Micalis Institute, Université Paris-Saclay, INRAE, AgroParisTech, 78350 Jouy-en-Josas, France; [email protected] (S.D.B.); [email protected] (M.M.); [email protected] (C.P.); [email protected] (H.M.B.) 2 Université Paris-Saclay, INRAE, MetaGenoPolis, 78350 Jouy-en-Josas, France 3 Enterome, 75011 Paris, France; [email protected] (W.F.); [email protected] (F.S.); [email protected] (J.-M.P.); [email protected] (L.C.) 4 INSERM UMRS 1138, Centre de Recherche des Cordeliers, Hôpital Pitié-Salpêtrière, Sorbonne-Université, 75006 Paris, France; [email protected] * Correspondence: [email protected]; Tel.: +33-134652428 Abstract: Non-alcoholic fatty liver diseases (NAFLD) are associated with changes in the composition and metabolic activities of the gut microbiota. However, the causal role played by the gut microbiota in individual susceptibility to NAFLD and particularly at its early stage is still unclear. In this context, we transplanted the microbiota from a patient with fatty liver (NAFL) and from a healthy individual to two groups of mice. We first showed that the microbiota composition in recipient mice Citation: Burz, S.D.; Monnoye, M.; resembled the microbiota composition of their respective human donor. Following administration Philippe, C.; Farin, W.; Ratziu, V.; Strozzi, F.; Paillarse, J.-M.; Chêne, L.; of a high-fructose, high-fat diet, mice that received the human NAFL microbiota (NAFLR) gained Blottière, H.M.; Gérard, P. -

Co-Infection Associated with Diarrhea in a Colony of <I>Scid
Laboratory Animal Science Vol 48, No 5 Copyright 1998 October 1998 by the American Association for Laboratory Animal Science Helicobacter bilis/Helicobacter rodentium Co-Infection Associated with Diarrhea in a Colony of scid Mice Nirah H. Shomer,* Charles A. Dangler, Robert P. Marini, and James G. Fox† Abstract _ An outbreak of diarrhea spanning 3 months occurred in a breeding colony of scid/Trp53 knockout mice. Approximately a third of the 150 mice were clinically affected, with signs ranging from mucoid or watery diarrhea to severe hemorrhagic diarrhea with mortality. Helicobacter bilis and the newly recognized urease-negative organ- ism H. rodentium were isolated from microaerobic culture of feces or cecal specimens from affected mice. Dual infection with H. bilis and H. rodentium were confirmed by culture and polymerase chain reaction (PCR) in several animals. Both Helicobacter species rapidly colonized immunocompetent sentinel mice exposed to bedding from cages containing affected mice, but the sentinel remained asymptomatic. Mice with diarrhea had multifocal to segmental proliferative typhlitis, colitis, and proctitis. Several affected mice had multifocal mucosal necrosis with a few focal ulcers in the cecum, colon, and rectum. Mice with diarrhea were treated with antibiotic food wafers (1.5 mg of amoxicillin, 0.69 mg of metronidazole, and 0.185 mg of bismuth/mouse per day) previously shown to eradi- cate H. hepaticus in immunocompetent mice. Antibiotic treatment resulted in resolution of diarrhea, but not eradication of H. bilis and H. rodentium; mice continued to have positive PCR results after a 2-week treatment regimen, and clinical signs of diarrhea returned in some mice when treatment was suspended. -

Enterohepatic Lesions in SCID Mice Infected with Helicobacter Bilis
Laboratory Animal Science Vol 48, No 4 Copyright 1998 August 1998 by the American Association for Laboratory Animal Science Enterohepatic Lesions in SCID Mice Infected with Helicobacter bilis Craig L. Franklin, Lela K. Riley, Robert S. Livingston, Catherine S. Beckwith, Cynthia L. Besch-Williford, and Reuel R. Hook, Jr. Abstract _ Helicobacter bilis is a recently identified species that colonizes the intestine and liver of mice. In immunocompetent mice, infections have been associated with mild hepatitis, and in immunocompromised mice, inflammatory bowel disease has been induced by intraperitoneal inoculation of the organism. We re- port inoculation of 6-week-old C.B-17 scid/scid mice by gastric gavage with approximately 107 H. bilis colony- forming units. Groups of mice were euthanized and necropsied 12, 24, and 36 weeks after inoculation. Mild to moderate proliferative typhlitis was evident in all mice at 12 and 36 weeks after inoculation and in most mice 24 weeks after inoculation. Mild to severe chronic active hepatitis was detected in 10 of 10 male mice and 3 of 10 female mice. These results indicate that H. bilis can cause moderate to severe enterohepatic disease in immunocompromised mice. The genus Helicobacter is a rapidly expanding genus volved in lesion development. Culture of specimens from currently containing 17 named species. Members of this mice confirmed intestinal colonization with H. hepaticus. Fox genus are microaerophilic, have curved to spiral rod mor- et al. reported enteric lesions in immunocompetent germ- phology, and are motile by flagella that vary in number free Swiss Webster mice infected with H. hepaticus (15), and and location among various species (1). -

Detection of Helicobacter Pylori in Faeces by Culture, PCR and Enzyme Immunoassay
J. Med. Microbiol. Ð Vol. 50 #2001), 1021±1029 # 2001 The Pathological Society of Great Britain and Ireland ISSN 0022-2615 REVIEW ARTICLE Detection of Helicobacter pylori in faeces by culture, PCR and enzyme immunoassay S. KABIR Academic Research and Information Management, 117 36 Stockholm, Sweden Various techniques such as culture, PCR and enzyme immunoassay have been used to detect Helicobacter pylori infection in human faecal specimens. Attempts to culture H. pylori have had limited success as the bacterium exists predominantly in a non- culturable coccoid)form in the faeces. Several PCR protocols, differing from each other in the choice of genomic targets and primers, have been used to detect H. pylori infection. Substances in faeces that inhibit PCR have been removed by various pre-PCR steps such as ®ltration through a polypropylene membrane, biochemical separation by column chromatography and isolation of H. pylori with immunomagnetic beads, the former two techniques yielding results with a high degree of sensitivity and speci®city. An enzyme immunoassay based on the detection of H. pylori antigen in faeces has become a convenient tool for the pre-treatment diagnosis of the infection. The stool antigen assay is convenient, especially for children, as it involves neither surgery nor the discomfort associated with the urea breath test. However, its applicability in monitoring eradication therapy has been controversial, as the assay can detect dead or partially degraded bacteria long after actual eradication, thus giving false positive results. Introduction faeces is compatible with a faecal±oral route of transmission, as faeces can contaminate the natural Helicobacter pylori is a fastidious, gram-negative, ¯ag- water supplies commonly used by people in poorer ellate bacterium known to colonise only man [1]. -

HEPATITIS a (Viral Or Infectious Hepatitis)
FACT SHEET HEPATITIS A (viral or infectious hepatitis) What is hepatitis A? Hepatitis A is a liver disease caused by hepatitis A virus. In children it may be very mild, but some adults who develop hepatitis A are ill enough to miss about four to six weeks of work. Who gets hepatitis A? Anyone can get hepatitis A, however, individuals who travel to countries where hepatitis A is common, intimate and household contacts of infected individuals, men who have sex with men and those who use illegal drugs are at an increased risk of becoming infected. How soon do symptoms appear? Time from infection to illness is 15 - 50 days with an average of 28 - 30 days. How is the virus spread? The hepatitis A virus is found in the feces (stool) of infected persons. It is usually spread by putting something in your mouth that has been contaminated by the stool of a person infected with hepatitis A. Hepatitis A may be spread by food that has been handled by infected persons who do not wash their hands carefully. Hepatitis A may also be spread by drinking water contaminated with human feces and the sharing of contaminated drug paraphernalia. What are the symptoms of hepatitis A? Fever, loss of appetite, nausea, vomiting, abdominal pains, and a general feeling of being ill are usually the first symptoms. These symptoms are typically followed in a few days by dark ("tea-colored") urine and jaundice (yellowing of the skin and the whites of the eyes). Infected persons usually feel better after one to two weeks, although they may continue to feel tired for a few more weeks. -

ACG Clinical Guideline: Management of Irritable Bowel Syndrome
CLINICAL GUIDELINES 17 ACG Clinical Guideline: Management of Irritable Bowel Syndrome Brian E. Lacy, PhD, MD, FACG1, Mark Pimentel, MD, FACG2, Darren M. Brenner, MD, FACG3, William D. Chey, MD, FACG4, 5 6 7 02/05/2021 on BhDMf5ePHKav1zEoum1tQfN4a+kJLhEZgbsIHo4XMi0hCywCX1AWnYQp/IlQrHD3i3D0OdRyi7TvSFl4Cf3VC4/OAVpDDa8K2+Ya6H515kE= by http://journals.lww.com/ajg from Downloaded Laurie A. Keefer, PhD , Millie D. Long, MDMPH, FACG (GRADE Methodologist) and Baha Moshiree, MD, MSc, FACG Downloaded Irritable bowel syndrome (IBS) is a highly prevalent, chronic disorder that significantly reduces patients’ quality of life. Advances in diagnostic testing and in therapeutic options for patients with IBS led to the development of this first-ever from http://journals.lww.com/ajg American College of Gastroenterology clinical guideline for the management of IBS using Grading of Recommendations, Assessment, Development, and Evaluation (GRADE) methodology. Twenty-five clinically important questions were assessed after a comprehensive literature search; 9 questions focused on diagnostic testing; 16 questions focused on therapeutic options. Consensus was obtained using a modified Delphi approach, and based on GRADE methodology, we endorse the by following: We suggest that a positive diagnostic strategy as compared to a diagnostic strategy of exclusion be used to improve BhDMf5ePHKav1zEoum1tQfN4a+kJLhEZgbsIHo4XMi0hCywCX1AWnYQp/IlQrHD3i3D0OdRyi7TvSFl4Cf3VC4/OAVpDDa8K2+Ya6H515kE= time to initiating appropriate therapy. We suggest that serologic testing be performed to rule out celiac disease in patients with IBS and diarrhea symptoms. We suggest that fecal calprotectin be checked in patients with suspected IBS and diarrhea symptoms to rule out inflammatory bowel disease. We recommend a limited trial of a low fermentable oligosaccharides, disacchardies, monosaccharides, polyols (FODMAP) diet in patients with IBS to improve global symptoms. -
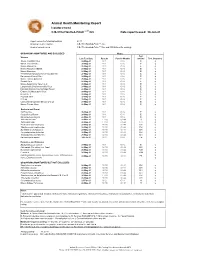
2021 ARC Health Report
Animal Health Monitoring Report Isolator reared C.B-17/IcrHanHsd-Prkdc scid /Arc Date report issued: 30-Jun-21 Report covers the following isolators: RI 27 Strains present in isolator: C.B-17/IcrHanHsd-Prkdc scid /Arc Strain of animal tested: C.B-17/IcrHanHsd-Prkdc scid /Arc and ICR Outbred for serology ORGANISMS MONITORED AND EXCLUDED Mouse Test Viruses Last Test Date Results Past 18 Months method Test frequency Mouse Hepatitis Virus 26-May-21 0 / 1 0 / 6 E q Minute Virus of Mice 26-May-21 0 / 1 0 / 6 E q Mouse Parvovirus 26-May-21 0 / 1 0 / 6 E q Murine Rotavirus (EDIM) 26-May-21 0 / 1 0 / 6 E q Mouse Norovirus 26-May-21 0 / 1 0 / 6 E q Theiler's Encephalomyelitis Virus (GD VII) 26-May-21 0 / 1 0 / 6 E q Pneumonia Virus of Mice 26-May-21 0 / 1 0 / 6 E q Murine Cytomegalovirus 26-May-21 0 / 1 0 / 6 E q Sendai Virus 26-May-21 0 / 1 0 / 6 E q Mouse Adenovirus Type 1 & 2 26-May-21 0 / 1 0 / 6 E q Lymphocytic Choriomeningitis Virus 26-May-21 0 / 1 0 / 6 E q Hantaan (Korean Haemorrhagic Fever) 26-May-21 0 / 1 0 / 6 E q Ectromelia (Mousepox) Virus 26-May-21 0 / 1 0 / 6 E q Reovirus -3 26-May-21 0 / 1 0 / 6 E q Polyoma Virus 26-May-21 0 / 1 0 / 6 E q K Virus 26-May-21 0 / 1 0 / 6 E q Lactic Dehydrogenase Elevating Virus 26-May-21 0 / 1 0 / 6 E q Mouse Thymic Virus 26-May-21 0 / 1 0 / 6 E q 00-Jan-00 Bacteria and Fungi CAR bacillus 26-May-21 0 / 1 0 / 6 E q Clostridium piliforme 26-May-21 0 / 1 0 / 6 E q Mycoplasma pulmonis 26-May-21 0 / 1 0 / 6 E q Helicobacter spp.1 26-May-21 0 / 10 0 / 44 H q Salmonella spp. -
R Graphics Output
883 | Desulfovibrio vulgaris | DvMF_2825 298701 | Desulfovibrio | DA2_3337 1121434 | Halodesulfovibrio aestuarii | AULY01000007_gene1045 207559 | Desulfovibrio alaskensis | Dde_0991 935942 | Desulfonatronum lacustre | KI912608_gene2193 159290 | Desulfonatronum | JPIK01000018_gene1259 1121448 | Desulfovibrio gigas | DGI_0655 1121445 | Desulfovibrio desulfuricans | ATUZ01000018_gene2316 525146 | Desulfovibrio desulfuricans | Ddes_0159 665942 | Desulfovibrio | HMPREF1022_02168 457398 | Desulfovibrio | HMPREF0326_00453 363253 | Lawsonia intracellularis | LI0397 882 | Desulfovibrio vulgaris | DVU_0784 1121413 | Desulfonatronovibrio hydrogenovorans | JMKT01000008_gene1463 555779 | Desulfonatronospira thiodismutans | Dthio_PD0935 690850 | Desulfovibrio africanus | Desaf_1578 643562 | Pseudodesulfovibrio aespoeensis | Daes_3115 1322246 | Pseudodesulfovibrio piezophilus | BN4_12523 641491 | Desulfovibrio desulfuricans | DND132_2573 1121440 | Desulfovibrio aminophilus | AUMA01000002_gene2198 1121456 | Desulfovibrio longus | ATVA01000018_gene290 526222 | Desulfovibrio salexigens | Desal_3460 1121451 | Desulfovibrio hydrothermalis | DESAM_21057 1121447 | Desulfovibrio frigidus | JONL01000008_gene3531 1121441 | Desulfovibrio bastinii | AUCX01000006_gene918 1121439 | Desulfovibrio alkalitolerans | dsat_0220 941449 | Desulfovibrio | dsx2_0067 1307759 | Desulfovibrio | JOMJ01000003_gene2163 1121406 | Desulfocurvus vexinensis | JAEX01000012_gene687 1304872 | Desulfovibrio magneticus | JAGC01000003_gene2904 573370 | Desulfovibrio magneticus | DMR_04750 -

Hepatitis a Summary and Frequently Asked Questions Updated 10/25/2017
Hepatitis A Summary and Frequently Asked Questions Updated 10/25/2017 Summary of San Diego hepatitis A Outbreak, 2017 The San Diego County Public Health Officer declared a local public health emergency on September 1, 2017 due to the ongoing hepatitis A virus outbreak in the county. The County Board of Supervisors ratified this declaration on September 6, 2017 and again on September 12, 2017. The County Board of Supervisors shall ratify the declaration every two weeks until the declaration is rescinded. Since early 2017, the County of San Diego Health and Human Services Agency’ Public Health Services has been investigating the hepatitis A outbreak. Control of the outbreak has been challenging because of the long time that it takes for the disease to develop (15 to 50 days) after a person is exposed to the infection (i.e. incubation period), and the difficulty of contacting many individuals sickened with the illness because they are homeless and/or illegal drug users. The outbreak is being spread person-to-person through contact with a fecal-contaminated environment. The majority of people who have contracted hepatitis A during this outbreak have been homeless and/or illegal drug users. This is not a foodborne outbreak; no common sources of food, beverage or drugs have been identified to contribute to this outbreak. The investigation, however, is ongoing. According to the Centers for Disease Control and Prevention (CDC), person-to-person transmission through close contact is the primary way people get hepatitis A in the United States1. Vaccination efforts are being implemented in targeted locations by County staff and, in collaboration with, health care partners. -

Helicobacter Pylori in Thai Patients with Cholangiocarcinoma and Its Association with Biliary Inflammation and Proliferation
DOI:10.1111/j.1477-2574.2011.00423.x HPB ORIGINAL ARTICLE Helicobacter pylori in Thai patients with cholangiocarcinoma and its association with biliary inflammation and proliferation Wongwarut Boonyanugomol1,5, Chariya Chomvarin1,5, Banchob Sripa2,5, Vajarabhongsa Bhudhisawasdi3,5, Narong Khuntikeo3,5, Chariya Hahnvajanawong1,5 & Amporn Chamsuwan4 1Department of Microbiology, 2Department of Pathology, 3Department of Surgery, 4Department of Forensic Medicine and 5Liver Fluke and Cholangiocarcinoma Research Center, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand Abstracthpb_423 177..184 Objectives: To investigate whether Helicobacter spp. infection and the cagA of H. pylori are associated with hepatobiliary pathology, specifically biliary inflammation, cell proliferation and cholangiocarcinoma (CCA). Methods: Helicobacter species including H. pylori, H. bilis and H. hepaticus were detected in the speci- mens using the polymerase chain reaction (PCR). Biliary inflammation of the liver and gallbladders was semi-quantitatively graded on hematoxylin and eosin (H&E)-stained slides. Biliary proliferation was evaluated by immunohistochemistry using the Ki-67-labelling index. Results: Helicobacter pylori was found in 66.7%, 41.5% and 25.0% of the patients in the CCA, cholelithiasis and control groups (P < 0.05), respectively. By comparison, H. bilis was found in 14.9% and 9.4% of the patients with CCA and cholelithiasis, respectively (P > 0.05), and was absent in the control group. The cagA gene of H. pylori was detected in 36.2% and 9.1% of the patients with CCA and cholelithiasis, respectively (P < 0.05). Among patients with CCA, cell inflammation and proliferation in the liver and gallbladder were significantly higher among those DNA H.