TITLE: Identification of Atlantic Cod (Gadus Morhua), Ling (Molva Molva) and Alaska Pollock
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Published Estimates of Life History Traits for 84 Populations of Teleost
Summary of data on fishing pressure group (G), age at maturity (Tm, years), length at maturity (Lm, cm), length-at-5%-survival (L.05, cm), time-to-5%-survival 3 (T.05, years), slope of the log-log fecundity-length relationship (Fb), fecundity the year of maturity (Fm), and egg volume (Egg, mm ) for the populations listed in the first three columns. Period is the period of field data collection. Species Zone Period G Tm Lm L.05 T.05 Fb Fm Egg Data sources (1) (1) (2) (3) (4) (4) (5) (1) (2) (3) (4) (5) Clupeiformes Engraulis capensis S. Africa 71-74 2 1 9.5 11.8 1.8 3.411 4.856E+04 0.988 118 119 137 118 138 Engraulis encrasicholus B. Biscay 87-92 2 1 11.5 14 1.4 3.997 9.100E+04 1.462 125 30, 188 170, 169 133, 23 145 Medit. S. 84-90 1 1 12.5 13.4 2.3 4.558 9.738E+04 0.668 161 161 160 161, 120 120 Sprattus sprattus Baltic S. 85-91 1 2 12 13.8 6.2 2.84 2.428E+05 1.122 15 19 26 184, 5 146 North S. 73-77 1 2 11.5 14.3 3 4.673 8.848E+03 0.393 8 107 106 33 169 Clupea harengus Baltic S. 75-82 1 3 16 24 4.9 3.206 4.168E+04 0.679 116 191 191 116 169 North S. 60-69 3 3 22 26.9 2.7 4.61 2.040E+04 0.679 52 53, 7 52 39 169 Baltic S. -

Histological Aspects of the Early Development of the Digestive System of Burbot Lota Lota L
ISSN 0015-5497, e-ISSN 1734-9168 Folia Biologica (Kraków), vol. 64 (2016), No 1 Ó Institute of Systematics and Evolution of Animals, PAS, Kraków, 2015 doi:10.3409/fb64_1.11 Histological Aspects of the Early Development of the Digestive System of Burbot Lota lota L. (Lotidae, Gadiformes)* Gra¿yna FURGA£A-SELEZNIOW, Ma³gorzata JANKUN, Roman KUJAWA, Joanna NOWOSAD, Maria BI£AS, Dariusz KUCHARCZYK, and Andrzej SKRZYPCZAK Accepted December 04, 2015 Published January 29, 2016 FURGA£A-SELEZNIOW G., JANKUN M., KUJAWA R., NOWOSAD J., BI£AS M., KUCHARCZYK D., SKRZYPCZAK A. 2016. Histological aspects of the early development of the digestive system of burbot Lota lota L. (Lotidae, Gadiformes). Folia Biologica (Kraków) 64: 11-21. The ontogeny of the digestive tract was studied histologically in burbot, Lota lota L., from hatching to 42 days post-hatch (dph). At hatching, the digestive tract consisted of a straight tube with discernible digestive accessory glands (the liver and the pancreas) dorsally attached to the yolk sac. Most of the yolk sac reserves were consumed during the first 12 days and were completely depleted by 17 dph. The first PAS-positive goblet cells appeared at 6 dph, dispersed within the epithelium of the oesophagus and increasing substantially in number and distribution as development progressed. At 12 dph, the first vacuoles (neutral lipids) appeared in the intestine, indicating the functional absorption of nutrients from food. Differentiation of gastric glands was first noticed at 17 dph and was extensive by 27 dph. L. lota larvae have a morphologically complete digestive tract by 32 dph. -

Scenario Calculations of Mercury Exposure
VKM Report 2019:3 Scenario calculations of mercury exposure from fish and overview of species with high mercury concentrations Opinion of the Panel on Contaminants of the Norwegian Scientific Committee for Food and Environment Report from the Norwegian Scientific Committee for Food and Environment (VKM) 2019:3 Scenario calculations of mercury exposure from fish and overview of species with high mercury concentrations Opinion of the Panel on Contaminants of the Norwegian Scientific Committee for Food and Environment 05.04.2019 ISBN: 978-82-8259-319-9 ISSN: 2535-4019 Norwegian Scientific Committee for Food and Environment (VKM) Po 4404 Nydalen N – 0403 Oslo Norway Phone: +47 21 62 28 00 Email: [email protected] vkm.no vkm.no/english Cover photo: Colourbox Suggested citation: VKM, Heidi Amlund, Kirsten Eline Rakkestad, Anders Ruus, Jostein Starrfelt, Jonny Beyer, Anne Lise Brantsæter, Sara Bremer, Gunnar Sundstøl Eriksen, Espen Mariussen, Ingunn Anita Samdal, Cathrine Thomsen and Helle Katrine Knutsen (2019). Scenario calculations of mercury exposure from fish and overview of species with high mercury concentrations. Opinion of the Panel on Contaminants of the Norwegian Scientific Committee for Food and Environment. VKM report 2019:3, ISBN: 978-82-8259-319-9, ISSN: 2535-4019. Norwegian Scientific Committee for Food and Environment (VKM), Oslo, Norway. Scenario calculations of mercury exposure from fish and overview of species with high mercury concentrations Preparation of the opinion The Norwegian Scientific Committee for Food and Environment (Vitenskapskomiteen for mat og miljø, VKM) appointed a project group to answer the request from the Norwegian Food Safety Authority. The project group consisted of three VKM-members, and three employees, including a project leader, from the VKM secretariat. -

9.2 Greater North Sea Ecoregion – Fisheries Overview
ICES Fisheries Overviews Greater North Sea Ecoregion Published 4 July 2017 DOI: 10.17895/ices.pub.3116 9.2 Greater North Sea Ecoregion – Fisheries overview Executive summary Around 6600 fishing vessels are active in the Greater North Sea. Total landings peaked in the 1970s at 4 million tonnes and have since declined to about 2 million tonnes. Total fishing effort has declined substantially since 2003. Pelagic fish landings are greater than demersal fish landings. Herring and mackerel, caught using pelagic trawls and seines, account for the largest portion of the pelagic landings, while sandeel and haddock, caught using otter trawls/seines, account for the largest fraction of the demersal landings. Catches are taken from more than 100 stocks. Discards are highest in the demersal and benthic fisheries. The spatial distribution of fishing gear varies across the Greater North Sea. Static gear is used most frequently in the English Channel, the eastern part of the Southern Bight, the Danish banks, and in the waters east of Shetland. Bottom trawls are used throughout the North Sea, with lower use in the shallower southern North Sea where beam trawls are most commonly used. Pelagic gears are used throughout the North Sea. In terms of tonnage of catch, most of the fish stocks harvested from the North Sea are being fished at levels consistent with achieving good environmental status (GES) under the EU’s Marine Strategy Framework Directive; however, the reproductive capacity of the stocks has not generally reached this level. Almost all the fisheries in the North Sea catch more than one species; controlling fishing on one species therefore affects other species as well. -

Marine Fishes from Galicia (NW Spain): an Updated Checklist
1 2 Marine fishes from Galicia (NW Spain): an updated checklist 3 4 5 RAFAEL BAÑON1, DAVID VILLEGAS-RÍOS2, ALBERTO SERRANO3, 6 GONZALO MUCIENTES2,4 & JUAN CARLOS ARRONTE3 7 8 9 10 1 Servizo de Planificación, Dirección Xeral de Recursos Mariños, Consellería de Pesca 11 e Asuntos Marítimos, Rúa do Valiño 63-65, 15703 Santiago de Compostela, Spain. E- 12 mail: [email protected] 13 2 CSIC. Instituto de Investigaciones Marinas. Eduardo Cabello 6, 36208 Vigo 14 (Pontevedra), Spain. E-mail: [email protected] (D. V-R); [email protected] 15 (G.M.). 16 3 Instituto Español de Oceanografía, C.O. de Santander, Santander, Spain. E-mail: 17 [email protected] (A.S); [email protected] (J.-C. A). 18 4Centro Tecnológico del Mar, CETMAR. Eduardo Cabello s.n., 36208. Vigo 19 (Pontevedra), Spain. 20 21 Abstract 22 23 An annotated checklist of the marine fishes from Galician waters is presented. The list 24 is based on historical literature records and new revisions. The ichthyofauna list is 25 composed by 397 species very diversified in 2 superclass, 3 class, 35 orders, 139 1 1 families and 288 genus. The order Perciformes is the most diverse one with 37 families, 2 91 genus and 135 species. Gobiidae (19 species) and Sparidae (19 species) are the 3 richest families. Biogeographically, the Lusitanian group includes 203 species (51.1%), 4 followed by 149 species of the Atlantic (37.5%), then 28 of the Boreal (7.1%), and 17 5 of the African (4.3%) groups. We have recognized 41 new records, and 3 other records 6 have been identified as doubtful. -
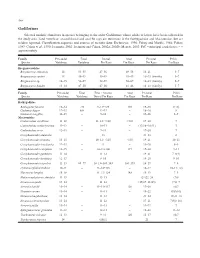
Gadiformes Selected Meristic Characters in Species Belonging to the Order Gadiformes Whose Adults Or Larvae Have Been Collected in the Study Area
548 Gadiformes Selected meristic characters in species belonging to the order Gadiformes whose adults or larvae have been collected in the study area. Total vertebrae, second dorsal and anal fin rays are numerous in the Bathygadidae and Macrouridae, but are seldom reported. Classification sequence and sources of meristic data: Eschmeyer, 1990; Fahay and Markle, 1984; Fahay, 1989; Cohen et al., 1990; Iwamoto, 2002; Iwamoto and Cohen, 2002a; 2002b; Merrett, 2003. PrC = principal caudal rays; ~ = approximately Family Precaudal Total Dorsal Anal Pectoral Pelvic Species Vertebrae Vertebrae Fin Rays Fin Rays Fin Rays Fin Rays Bregmacerotidae Bregmaceros atlanticus 14 53–55 47–56 49–58 16–21 5–7 Bregmaceros cantori 14 45–49 45–49 45–49 16–23 (family) 5–7 Bregmaceros sp. 14–15 52–59 52–59 58–69 16–23 (family) 5–7 Bregmaceros houdei 13–14 47–50 47–50 41–46 16–23 (family) 5–7 Family Precaudal Total First + Second Anal Pectoral Pelvic Species Vertebrae Vertebrae Dorsal Fin Rays Fin Rays Fin Rays Fin Rays Bathygadidae Bathygadus favosus 12–14 ~70 9–11+125 110 15–18 9(10) Gadomus dispar 12–13 80+ 12–13 – 18–20 8 Gadomus longifilis 11–13 – 9–11 – 14–16 8–9 Macrouridae Caelorinchus caribbeus 11–12 – 11–12+>110 >110 17–20 7 Caelorinchus coelorhynchus 11–12 – 10–11 – (17)18–20(21) 7 Caelorinchus occa 12–13 – 9–11 – 17–20 7 Coryphaenoides alateralis – 13 – 21–23 8 Coryphaenoides armatus 13–15 – 10–12+~125 ~135 19–21 10–11 Coryphaenoides brevibarbis 12–13 – 9 – 19–20 8–9 Coryphaenoides carapinus 12–15 – 10–11+100 117 17–20 9–11 Coryphaenoides guentheri -

Intrinsic Vulnerability in the Global Fish Catch
The following appendix accompanies the article Intrinsic vulnerability in the global fish catch William W. L. Cheung1,*, Reg Watson1, Telmo Morato1,2, Tony J. Pitcher1, Daniel Pauly1 1Fisheries Centre, The University of British Columbia, Aquatic Ecosystems Research Laboratory (AERL), 2202 Main Mall, Vancouver, British Columbia V6T 1Z4, Canada 2Departamento de Oceanografia e Pescas, Universidade dos Açores, 9901-862 Horta, Portugal *Email: [email protected] Marine Ecology Progress Series 333:1–12 (2007) Appendix 1. Intrinsic vulnerability index of fish taxa represented in the global catch, based on the Sea Around Us database (www.seaaroundus.org) Taxonomic Intrinsic level Taxon Common name vulnerability Family Pristidae Sawfishes 88 Squatinidae Angel sharks 80 Anarhichadidae Wolffishes 78 Carcharhinidae Requiem sharks 77 Sphyrnidae Hammerhead, bonnethead, scoophead shark 77 Macrouridae Grenadiers or rattails 75 Rajidae Skates 72 Alepocephalidae Slickheads 71 Lophiidae Goosefishes 70 Torpedinidae Electric rays 68 Belonidae Needlefishes 67 Emmelichthyidae Rovers 66 Nototheniidae Cod icefishes 65 Ophidiidae Cusk-eels 65 Trachichthyidae Slimeheads 64 Channichthyidae Crocodile icefishes 63 Myliobatidae Eagle and manta rays 63 Squalidae Dogfish sharks 62 Congridae Conger and garden eels 60 Serranidae Sea basses: groupers and fairy basslets 60 Exocoetidae Flyingfishes 59 Malacanthidae Tilefishes 58 Scorpaenidae Scorpionfishes or rockfishes 58 Polynemidae Threadfins 56 Triakidae Houndsharks 56 Istiophoridae Billfishes 55 Petromyzontidae -

The Complete Mitochondrial Genome of the Whiting, Merlangius
CORE Metadata, citation and similar papers at core.ac.uk Provided by Digital.CSIC The complete mitochondrial genome of the whiting, Merlangius merlangus and the haddock, Melanogrammus aeglefinus: A detailed genomic comparison among closely related species of the Gadidae family Severine Roques a , Clive J. Fox b , Maria I. Villasana a , Ciro Rico a,⁎ a Estación Biológica de Doñana, CSIC, Pabellón del Perú, 41013 Sevilla, Spain b The Centre for Environment, Fisheries and Aquaculture Science, Pakefield Road, Lowestoft, Suffolk, NR33 OHT, England, United Kingdom Abstract We determined the first complete mitochondrial DNA (mtDNA) sequences for the whiting (Merlangius merlangus, family Gadidae, order Gadiformes) and the haddock (Melanogrammus aeglefinus, family Gadidae, order Gadiformes). The entire mitogenomes were amplified and sequenced by primer walking using newly designed specific internal primers. Lengths were 16,569 and 16,585 bases for whiting and haddock respectively, lengths which lie within the range of previously reported gadiform sequences from Atlantic cod (Gadus morhua, 16,696 bases) and walleye pollock (Theragra chalcogramma, 16,570 bases). Gene arrangement in both species conformed to the order seen in most vertebrate mitochondrial genomes. We identified a long intergenic spacer located between the tRNAThr and tRNAPro genes (of 100 and 70 bp long for whiting and haddock, respectively), as previously described for other species of the order Gadiformes. Using nucleotide and amino acid divergence data of four complete gadoid mitogenomes (M. merlangius, M. aeglefinus, G. morhua and T. chalcogramma), we examined in detail the relative mtDNA mutation patterns across genes and among Gadidae species and tested for the performance of each protein-coding, transfer RNA and ribosomal RNA gene in depicting the expected phylogeny among the four species, as compared with the whole genome dataset. -

European Trawlers Are Destroying the Oceans
EUROPEAN TRAWLERS ARE DESTROYING THE OCEANS Introduction Nearly 100,000 vessels make up the European Union fishing fleet. This includes boats that fish both in EU waters (the domestic fleet), in the waters of other countries and in international waters (the deep-sea fleet). In addition, there is an unknown number of vessels belonging to other European countries that are not members of the EU which could approach a figure half that of the EU fleet. The majority of these vessels sail under the flag of a European country but there are also boats, particularly those fishing on the high seas, which despite being managed, chartered or part owned by European companies, use the flag of the country where they catch their fish or sail under flags of convenience (FOCs). The Fisheries Commission has called for a reform of the Common Fisheries Policy (CFP) to achieve a reduction of 40% in the EU fishing capacity, as forecasts show that by simply following the approved multi-annual plans, barely 8.5% of vessels and 18% of gross tonnage would be decommissioned1; an achievement very distant from scientific recommendations. Moreover, from among these almost 100,000 vessels, the EU is home to a particularly damaging fleet: the 15,000 trawlers that operate in European waters, as well as those of third countries or those fishing on the high seas. These trawlers are overexploiting marine resources and irreversibly damaging some of the most productive and biodiverse ecosystems on the planet. The 40% reduction called for by the Commission could be easily achieved if the primary objective of this proposal was focused both on eliminating the most destructive fishing techniques and reducing fishing overcapacity. -

Length-Weight Relationships of Marine Fish Collected from Around the British Isles
Science Series Technical Report no. 150 Length-weight relationships of marine fish collected from around the British Isles J. F. Silva, J. R. Ellis and R. A. Ayers Science Series Technical Report no. 150 Length-weight relationships of marine fish collected from around the British Isles J. F. Silva, J. R. Ellis and R. A. Ayers This report should be cited as: Silva J. F., Ellis J. R. and Ayers R. A. 2013. Length-weight relationships of marine fish collected from around the British Isles. Sci. Ser. Tech. Rep., Cefas Lowestoft, 150: 109 pp. Additional copies can be obtained from Cefas by e-mailing a request to [email protected] or downloading from the Cefas website www.cefas.defra.gov.uk. © Crown copyright, 2013 This publication (excluding the logos) may be re-used free of charge in any format or medium for research for non-commercial purposes, private study or for internal circulation within an organisation. This is subject to it being re-used accurately and not used in a misleading context. The material must be acknowledged as Crown copyright and the title of the publication specified. This publication is also available at www.cefas.defra.gov.uk For any other use of this material please apply for a Click-Use Licence for core material at www.hmso.gov.uk/copyright/licences/ core/core_licence.htm, or by writing to: HMSO’s Licensing Division St Clements House 2-16 Colegate Norwich NR3 1BQ Fax: 01603 723000 E-mail: [email protected] 3 Contents Contents 1. Introduction 5 2. -

Establishment and Partial Characterization of a Cell Line from Burbot Lota Lota Maculosa: Susceptibility to IHNV, IPNV and VHSV Mark P
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln USGS Staff -- ubP lished Research US Geological Survey 5-18-2010 Establishment and partial characterization of a cell line from burbot Lota lota maculosa: susceptibility to IHNV, IPNV and VHSV Mark P. Polinski Department of Fish and Wildlife Resources, College of Natural Resources, University of Idaho John D. Drennan Intervet/Schering-Plough Animal Health William N. Batts US Geological Survey, Western Fisheries Research Center Susan C. Ireland Kootenai Tribe of Idaho Kenneth D. Cain Department of Fish and Wildlife Resources, College of Natural Resources, University of Idaho,, [email protected] Follow this and additional works at: http://digitalcommons.unl.edu/usgsstaffpub Part of the Geology Commons, Oceanography and Atmospheric Sciences and Meteorology Commons, Other Earth Sciences Commons, and the Other Environmental Sciences Commons Polinski, Mark P.; Drennan, John D.; Batts, iW lliam N.; Ireland, Susan C.; and Cain, Kenneth D., "Establishment and partial characterization of a cell line from burbot Lota lota maculosa: susceptibility to IHNV, IPNV and VHSV" (2010). USGS Staff -- Published Research. 1062. http://digitalcommons.unl.edu/usgsstaffpub/1062 This Article is brought to you for free and open access by the US Geological Survey at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in USGS Staff -- ubP lished Research by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Vol. 90: 15–23, 2010 DISEASES OF AQUATIC ORGANISMS Published May 18 doi: 10.3354/dao02215 Dis Aquat Org Establishment and partial characterization of a cell line from burbot Lota lota maculosa: susceptibility to IHNV, IPNV and VHSV Mark P. -

Comparison of Size Selectivity Between Marine Mammals and Commercial Fisheries with Recommendations for Restructuring Management Policies
NOAA Technical Memorandum NMFS-AFSC-159 Comparison of Size Selectivity Between Marine Mammals and Commercial Fisheries with Recommendations for Restructuring Management Policies by M. A. Etnier and C. W. Fowler U.S. DEPARTMENT OF COMMERCE National Oceanic and Atmospheric Administration National Marine Fisheries Service Alaska Fisheries Science Center October 2005 NOAA Technical Memorandum NMFS The National Marine Fisheries Service's Alaska Fisheries Science Center uses the NOAA Technical Memorandum series to issue informal scientific and technical publications when complete formal review and editorial processing are not appropriate or feasible. Documents within this series reflect sound professional work and may be referenced in the formal scientific and technical literature. The NMFS-AFSC Technical Memorandum series of the Alaska Fisheries Science Center continues the NMFS-F/NWC series established in 1970 by the Northwest Fisheries Center. The NMFS-NWFSC series is currently used by the Northwest Fisheries Science Center. This document should be cited as follows: Etnier, M. A., and C. W. Fowler. 2005. Comparison of size selectivity between marine mammals and commercial fisheries with recommendations for restructuring management policies. U.S. Dep. Commer., NOAA Tech. Memo. NMFS-AFSC-159, 274 p. Reference in this document to trade names does not imply endorsement by the National Marine Fisheries Service, NOAA. NOAA Technical Memorandum NMFS-AFSC-159 Comparison of Size Selectivity Between Marine Mammals and Commercial Fisheries with Recommendations for Restructuring Management Policies by M. A. Etnier and C. W. Fowler Alaska Fisheries Science Center 7600 Sand Point Way N.E. Seattle, WA 98115 www.afsc.noaa.gov U.S. DEPARTMENT OF COMMERCE Carlos M.