Hodgkin B Cell Lymphomas Identified by BH3 Profiling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Optimal Minimal Panels of Immunohistochemistry for Diagnosis of B-Cell Lymphoma for Application in Countries with Limited Resources and for Triaging Cases Before
AJCP /ORIGINAL ARTICLE Optimal Minimal Panels of Immunohistochemistry for Diagnosis of B-Cell Lymphoma for Application in Countries With Limited Resources and for Triaging Cases Before Referral to Specialist Centers Downloaded from https://academic.oup.com/ajcp/article-abstract/145/5/687/2195691 by World Health Organization user on 09 January 2019 Maria Giulia Disanto, MD,1 Maria Raffaella Ambrosio, MD, PhD,2 Bruno Jim Rocca, MD, PhD,2 Hazem A. H. Ibrahim, FRCPath, PhD,1,3 Lorenzo Leoncini, MD, PhD,2 and Kikkeri N. Naresh, MD, FRCPath1 From the 1Department of Histopathology, Imperial College Healthcare NHS Trust & Imperial College, London, United Kingdom; 2Department of Medical Biotechnologies, Section of Pathology, University of Siena, Siena, Italy; and 3Department of Histopathology, Faculty of Medicine, Mansoura University, Mansoura, Egypt. Key Words: Lymphoma; B-cell lymphoma; Immunohistochemistry; Diagnosis; Classification; Developing countries Am J Clin Pathol May 2016;145:687-695 DOI: 10.1093/AJCP/AQW060 ABSTRACT Lymphomas are a collection of different malignancies “arising” from lymphoid cells. They include about 49 entities, Objectives: Establish and validate optimal minimal and over 19 provisional entities and subsets.1 About 85% of immunohistochemistry panels for usage in a staged lymphomas are of B-cell origin. Precision in lymphoma diag- algorithmic manner for precise diagnosis of B-cell nosis requires expertise and infrastructure. The entities are lymphomas in countries with limited resources. Suggest defined based on morphology, immunohistochemistry (on short panels of immunostains to be used in referring units some occasions in situ hybridization), cytogenetics/fluores- that refer suspected lymphomas to specialist diagnostic cent in situ hybridization (FISH), molecular genetics and clin- centers in resourceful countries. -

RA0358-C.5-IFU-RUO CD57 / B3GAT1 (Natural Killer Cell
Instructions For Use RA0 35 8-C.5 -IFU -RUO Revision: 1 Rev. Date: Dec. 19, 2014 Page 1 of 2 P.O. Box 3286 - Logan, Utah 84323, U.S.A. - Tel. (800) 729-8350 – Tel. (435) 755-9848 - Fax (435) 755-0015 - www.scytek.com CD57 / B3GAT1 (Natural Killer Cell Marker); Clone NK/804 (Concentrate) Availability/Contents: Item # Volume RA0358-C.5 0.5 ml Description: Species: Mouse Immunogen: Recombinant human B3GAT1 protein Clone: NK/804 Isotype: IgM, kappa Entrez Gene ID: 27087 (Human) Hu Chromosome Loc.: 11q25 Synonyms: 3-Glucuronyltransferase 1; B3GAT1; Galactosylgalactosylxylosylprotein 3-beta- Glucuronosyltransferase 1; GLCATP; GlcUAT-P; Glucuronosyltransferase P; UDP GlcUA Glycoprotein beta 1, 3 Glucuronyltransferase. Mol. Weight of Antigen: ~110kDa Format: Bioreactor Concentrate with 0.05% Azide. Specificity: Anti-CD57 marks a subset of lymphocytes known as natural killer (NK) cells. Anti-CD57 also stains neuroendocrine cells and their derived tumors, including carcinoid tumors and medulloblastoma. Background: Follicular center cell lymphomas often contain many NK cells within the neoplastic follicles. Anti- CD57 can be useful in separating type B3 thymoma from thymic carcinoma when combined with a panel that includes antibodies against GLUT1, CD5, and CEA. Species Reactivity: Human. Does not react with Rat. Others not known. Positive Control: Lymph node or tonsil. Cellular Localization: Cell surface Titer/ Working Dilution: Immunohistochemistry (Frozen and Formalin-fixed): 1:50-1:100 Flow Cytometry: 5-10 µl/million cells Immunofluorescence: 1:50-1:100 Western Blotting: 1:100-1:200 Microbiological State: This product is not sterile. 8° C Storage: 2° C C ScyTek Laboratories, Inc. 205 South 600 West P EmergoEurope (31)(0) 70 345-8570 Logan, UT 84321 Molsnstraat 15 Doc: IFU-Template2-8rev2 U.S.A. -

Flow Cytometry CPT: 88182, 88184, 88185, 88187, 88188, 88189, 86355, 86356, 86357, 86359, 86360, 86361, 86367
Medicare Local Coverage Determination Policy Flow Cytometry CPT: 88182, 88184, 88185, 88187, 88188, 88189, 86355, 86356, 86357, 86359, 86360, 86361, 86367 CMS Policy for Alaska, Arizona, Idaho, Montana, North Dakota, Medically Supportive Oregon, South Dakota, Utah, Washington, and Wyoming ICD Codes are listed Local policies are determined by the performing test location. This is determined by the state on subsequent page(s) in which your performing laboratory resides and where your testing is commonly performed. of this document. Coverage Indications, Limitations, and/or Medical Necessity Flow cytometry (FCM) is a complex process to examine blood, body fluids, CSF, bone marrow, lymph node, tonsil, spleen and other solid tissues. The use of peripheral blood and fine needle aspirate material avoids more invasive procedures for diagnosis. A flow cytometer evaluates the physical and/or chemical characteristics of single cells as the cells pass individually in a fluid stream through a measuring device. Surface receptors, intracellular molecules, and DNA bind with fluorescent dyes that allow detection and evaluation. When light of one wave length excites electrons of certain chemicals to energy levels above their ground state and upon return to ground state emits light of a longer wavelength, fluorescence is produced. A flow cytometer detects cell characteristics by measuring the fluorescence produced by fluorochromes conjugated either directly with cell components or conjugated to antibodies directed against cell components. Indications • Cytopenias and Hypercellular Hematolymphoid Disorders Hematolymphoid neoplasia can present with cytopenias (anemia, leucopenia and/or thrombocytopenia) or elevated leukocyte counts. If medical review and preliminary laboratory testing fails to reveal a cause, bone marrow aspiration and biopsy are indicated to rule out an infiltrative process or a stem cell disorder. -

CD3+CD5+CD4-CD8-) Alpha/Beta T Cell Receptor-Bearing Cells
Mlsa generated suppressor cells. I. Suppression is mediated by double-negative (CD3+CD5+CD4-CD8-) alpha/beta T cell receptor-bearing cells. This information is current as of October 2, 2021. M Bruley-Rosset, I Miconnet, C Canon and O Halle-Pannenko J Immunol 1990; 145:4046-4052; ; http://www.jimmunol.org/content/145/12/4046 Downloaded from Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision http://www.jimmunol.org/ • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription by guest on October 2, 2021 Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 1990 by American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. 0022-1767/90/14512-4046$02.00/0 THEJOURNAL OF IMMUNOLCGY Vol. 145.4046-4052. No. 12. December 15. 1990 Copyright 0 1990 by The American Association of lmmunologists Printed In U.S.A. Mls" GENERATED SUPPRESSORCELLS I. Suppression is Mediated by Double-Negative (CD3+CDVCD4-CD8-) a/@ T Cell Receptor-Bearing Cells' Grafting of cells from B10.D2 (H-2d)donors into H- The GVHR3 remains a major problem after bone mar- 2 compatible lethally irradiated (DBA/2 x B1O.DZ)Fl row transplantation, even in MHC-compatible donor-re- hostsresults in a severe graft-vs-host reaction cipient combinations. -

Ligand for Bovine CD5 the Identification and Characterization of A
The Identification and Characterization of a Ligand for Bovine CD5 Karen M. Haas and D. Mark Estes This information is current as J Immunol 2001; 166:3158-3166; ; of September 27, 2021. doi: 10.4049/jimmunol.166.5.3158 http://www.jimmunol.org/content/166/5/3158 Downloaded from References This article cites 46 articles, 18 of which you can access for free at: http://www.jimmunol.org/content/166/5/3158.full#ref-list-1 Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on September 27, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2001 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Identification and Characterization of a Ligand for Bovine CD51 Karen M. Haas* and D. Mark Estes2*† CD5, a type I glycoprotein expressed by T cells and a subset of B cells, is thought to play a significant role in modulating Ag receptor signaling. Previously, our laboratory has shown that bovine B cells are induced to express this key regulatory molecule upon Ag receptor cross-linking. -

B-Cell Activation by Crosslinking of Surface Igm Or Ligation of CD40 Involves Alternative Signal Pathways and Results in Different B-Cell Phenotypes HENRY H
Proc. Natl. Acad. Sci. USA Vol. 92, pp. 3348-3352, April 1995 Immunology B-cell activation by crosslinking of surface IgM or ligation of CD40 involves alternative signal pathways and results in different B-cell phenotypes HENRY H. WORTIS*t, MARK TEUTSCH*, MINDY HIGER*, JENNY ZHENG*, AND DAVID C. PARKERt§ *Department of Pathology, Tufts University School of Medicine, and Graduate Program in Immunology, Sackler School of Graduate Biomedical Sciences, Boston, MA 02111; and tDepartment of Molecular Genetics and Microbiology, University of Massachusetts Medical School, Worcester, MA 01655 Communicated by Salome G. Waelsch, Albert Einstein College of Medicine of Yeshiva University, Bronx, NY December 7, 1994 ABSTRACT Treatment of small resting B cells with sol- direct contact with an activated T-helper cell such that the uble F(ab')2 fragments of anti-IgM, an analogue of T-inde- surface molecule gp39 [CD40 ligand (CD40L)] of the T cell pendent type 2 antigens, induced activation characterized by ligates the CD40 of the B cell (10). proliferation and the expression of surface CD5. In contrast, Our strategy to determine if minimal TD and TI-2 signals B cells induced to proliferate in response to thymus-dependent were sufficient to induce phenotypic differences in B cells was inductive signals provided by either fixed activated T-helper 2 to use an antigen that could be modified so as to initiate either cells or soluble CD40 ligand-CD8 (CD40L) recombinant pro- a TD or a TI-2 response. To produce a TD response we used tein displayed elevated levels of CD23 (FcJII receptor) and no monovalent Fab of rabbit anti-IgM and provided help in the surface CD5. -

Flow Cytometric Analysis of B-Cell Lymphoproliferative Disorders
Flow cytometric analysis of B-cell lymphoproliferative disorders David M. Dorfman, M.D., Ph.D. Department of Pathology Brigham and Women’s Hospital and Harvard Medical School Boston, MA Objectives • Review basic principles of flow cytometric immunophenotypic analysis of B cell lymphoproliferative disorders • Discuss recent studies to overcome limitations and shortcomings – New markers – New methods Incidence of B-cell neoplasms, United States Subtype Incidence rate 2011-2012 New cases, 2016 per 100,000 Lymphoid neoplasms 34.4 136,960 Lymphoid neoplasms, B 29.0 93.3% 117,470 B-LL/L 1.4 82.2% 4,930 CLL/SLL 5.1 20,980 FL 3.4 13,960 DLBCL 6.3 27,650 MM 5.9 24,280 Lymphoid neoplasms, T/NK 2.1 8,380 T-LL/L 0.3 1,070 T-PLL <0.1 160 T-LGL 0.2 670 ATL/L <0.1 180 Teras et al. CA Cancer J Clin 2016; 66:443-459 (North American AssociationTeras of et Central al. CA Cancer Cancer Registries) J Clin 2016; 66:443-459 (North American Association of Central Cancer Registries) SS <0.1 Teras et al. CA Cancer J Clin70 2016; 66:443-459 (North American Association of Central Cancer Registries) 94% WHO revised 4th ed., 2017 Flow cytometric analysis of B-cell lymphoproliferative disorders • B-cell antigen expression (CD19, CD20, CD22) • Monoclonal surface immunoglobulin κ or λ light chain expression (or absence of surface immunoglobulin) • Expression of additional B-cell antigens or other antigens, including abnormal expression levels • Presence of cells with abnormal light scatter characteristics ( high forward scatter or side scatter) B-ALL MCL FL, HL MZL, CLL, MM LPL DLBCL DLBCL WHO revised 4th ed. -

Functional Studies of Chronic Lymphocytic Leukemia B Cells Expressing Β2-Integrin Type Complement Receptors CR3 and CR4
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Repository of the Academy's Library Functional studies of chronic lymphocytic leukemia B cells expressing β2-integrin type complement receptors CR3 and CR4 Barbara Uzonyia,1, Bernadett Mácsik-Valentb,1, Szilvia Lukácsib, Richárd Kissc, Katalin Törökb, Mariann Kremlitzkaa,2, Zsuzsa Bajtayb, Judit Demeterd, Csaba Bödörc, Anna Erdeia,b,* a MTA-ELTE Immunology Research Group, Department of Immunology, Eötvös Loránd University, Budapest, Hungary b Department of Immunology, Eötvös Loránd University, Budapest, Hungary c MTA-SE “Lendület” Molecular Oncohematology Research Group, 1st Department of Pathology, and Experimental Cancer Research, Semmelweis University, Budapest, Hungary d 1st Department of Internal Medicine, Semmelweis University, Budapest, Hungary 1 These authors contributed equally to this work 2 Present address: Division of Medical Protein Chemistry, Department of Translational Medicine, Lund University, Malmö, Sweden * Corresponding author Anna Erdei Department of Immunology, Eötvös Loránd University, Pázmány Péter s. 1/C, Budapest H-1117, Hungary Phone: (+36)-1-3812-175 Fax: (+36)-1-3812-176 e-mail: [email protected], [email protected] Abbreviations BCR: B cell receptor, CLL: chronic lymphocytic leukemia, CR: complement receptor, IL: interleukin Keywords CLL B cells, IL-10 production, expression of CR3 and CR4, adhesion and spreading 1 Abstract The expression and role of CR3 (CD11b/CD18) and CR4 (CD11c/CD18) in B cells are not yet explored in contrast to myeloid cells, where these β2-integrin type receptors are known to participate in various cellular functions, including phagocytosis, adherence and migration. Here we aimed to reveal the expression and role of CR3 and CR4 in human B cells. -
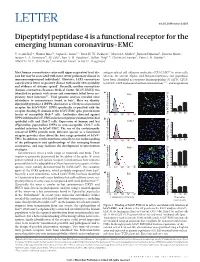
Dipeptidyl Peptidase 4 Is a Functional Receptor for the Emerging Human Coronavirus-EMC
LETTER doi:10.1038/nature12005 Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC V. Stalin Raj1*, Huihui Mou2*, Saskia L. Smits1,3, Dick H. W. Dekkers4, Marcel A. Mu¨ller5, Ronald Dijkman6, Doreen Muth5, Jeroen A. A. Demmers4, Ali Zaki7, Ron A. M. Fouchier1, Volker Thiel6,8, Christian Drosten5, Peter J. M. Rottier2, Albert D. M. E. Osterhaus1, Berend Jan Bosch2 & Bart L. Haagmans1 Most human coronaviruses cause mild upper respiratory tract dis- antigen-related cell adhesion molecules (CEACAM)10 to enter cells, ease but may be associated with more severe pulmonary disease in whereas for several Alpha- and Betacoronaviruses, two peptidases immunocompromised individuals1. However, SARS coronavirus have been identified as receptors (aminopeptidase N (APN, CD13) caused severe lower respiratory disease with nearly 10% mortality for hCoV-229E and several animal coronaviruses11,12, and angiotensin and evidence of systemic spread2. Recently, another coronavirus (human coronavirus-Erasmus Medical Center (hCoV-EMC)) was a identified in patients with severe and sometimes lethal lower res- )] Vero –1 piratory tract infection3,4. Viral genome analysis revealed close 9 ml 8 5 50 relatedness to coronaviruses found in bats . Here we identify 7 dipeptidyl peptidase 4 (DPP4; also known as CD26) as a functional 6 5 receptor for hCoV-EMC. DPP4 specifically co-purified with the 4 receptor-binding S1 domain of the hCoV-EMC spike protein from 3 Relative cell number log[GE (TCID 0 20 40 lysates of susceptible Huh-7 cells. Antibodies directed against 0 101 102 103 104 DPP4 inhibited hCoV-EMC infection of primary human bronchial b )] COS-7 –1 epithelial cells and Huh-7 cells. -

IL-21 in Conjunction with Anti-CD40 and IL-4 Constitutes a Potent Polyclonal B Cell Stimulator for Monitoring Antigen-Specific Memory B Cells
cells Article IL-21 in Conjunction with Anti-CD40 and IL-4 Constitutes a Potent Polyclonal B Cell Stimulator for Monitoring Antigen-Specific Memory B Cells Fridolin Franke 1,2, Greg A. Kirchenbaum 1 , Stefanie Kuerten 2 and Paul V. Lehmann 1,* 1 Research & Development Department, Cellular Technology Limited, Shaker Heights, OH 44122, USA; [email protected] (F.F.); [email protected] (G.A.K.) 2 Institute of Anatomy and Cell Biology, Friedrich-Alexander University Erlangen-Nürnberg, 91054 Erlangen, Germany; [email protected] * Correspondence: [email protected]; Tel.: +1-216-965-6311 Received: 3 December 2019; Accepted: 12 February 2020; Published: 13 February 2020 Abstract: Detection of antigen-specific memory B cells for immune monitoring requires their activation, and is commonly accomplished through stimulation with the TLR7/8 agonist R848 and IL-2. To this end, we evaluated whether addition of IL-21 would further enhance this TLR-driven stimulation approach; which it did not. More importantly, as most antigen-specific B cell responses are T cell-driven, we sought to devise a polyclonal B cell stimulation protocol that closely mimics T cell help. Herein, we report that the combination of agonistic anti-CD40, IL-4 and IL-21 affords polyclonal B cell stimulation that was comparable to R848 and IL-2 for detection of influenza-specific memory B cells. An additional advantage of anti-CD40, IL-4 and IL-21 stimulation is the selective activation of IgM+ memory B cells, as well as the elicitation of IgE+ ASC, which the former fails to do. Thereby, we introduce a protocol that mimics physiological B cell activation through helper T cells, including induction of all Ig classes, for immune monitoring of antigen-specific B cell memory. -

Human CD Marker Chart Reviewed by HLDA1 Bdbiosciences.Com/Cdmarkers
BD Biosciences Human CD Marker Chart Reviewed by HLDA1 bdbiosciences.com/cdmarkers 23-12399-01 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD1a R4, T6, Leu6, HTA1 b-2-Microglobulin, CD74 + + + – + – – – CD93 C1QR1,C1qRP, MXRA4, C1qR(P), Dj737e23.1, GR11 – – – – – + + – – + – CD220 Insulin receptor (INSR), IR Insulin, IGF-2 + + + + + + + + + Insulin-like growth factor 1 receptor (IGF1R), IGF-1R, type I IGF receptor (IGF-IR), CD1b R1, T6m Leu6 b-2-Microglobulin + + + – + – – – CD94 KLRD1, Kp43 HLA class I, NKG2-A, p39 + – + – – – – – – CD221 Insulin-like growth factor 1 (IGF-I), IGF-II, Insulin JTK13 + + + + + + + + + CD1c M241, R7, T6, Leu6, BDCA1 b-2-Microglobulin + + + – + – – – CD178, FASLG, APO-1, FAS, TNFRSF6, CD95L, APT1LG1, APT1, FAS1, FASTM, CD95 CD178 (Fas ligand) + + + + + – – IGF-II, TGF-b latency-associated peptide (LAP), Proliferin, Prorenin, Plasminogen, ALPS1A, TNFSF6, FASL Cation-independent mannose-6-phosphate receptor (M6P-R, CIM6PR, CIMPR, CI- CD1d R3G1, R3 b-2-Microglobulin, MHC II CD222 Leukemia