From Clone to Bone: the Synergy of Morphological and Molecular Tools in Palaeobiology Edited by Robert J
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evolution of the Patellar Sesamoid Bone in Mammals
A peer-reviewed version of this preprint was published in PeerJ on 21 March 2017. View the peer-reviewed version (peerj.com/articles/3103), which is the preferred citable publication unless you specifically need to cite this preprint. Samuels ME, Regnault S, Hutchinson JR. 2017. Evolution of the patellar sesamoid bone in mammals. PeerJ 5:e3103 https://doi.org/10.7717/peerj.3103 Evolution of the patellar sesamoid bone in mammals Mark E Samuels 1, 2 , Sophie Regnault 3 , John R Hutchinson Corresp. 3 1 Department of Medicine, University of Montreal, Montreal, Quebec, Canada 2 Centre de Recherche du CHU Ste-Justine, Montreal, Quebec, Canada 3 Structure & Motion Laboratory, Department of Comparative Biomedical Sciences, The Royal Veterinary College, Hatfield, Hertfordshire, United Kingdom Corresponding Author: John R Hutchinson Email address: [email protected] The patella is a sesamoid bone located in the major extensor tendon of the knee joint, in the hindlimb of many tetrapods. Although numerous aspects of knee morphology are ancient and conserved among most tetrapods, the evolutionary occurrence of an ossified patella is highly variable. Among extant (crown clade) groups it is found in most birds, most lizards, the monotreme mammals and almost all placental mammals, but it is absent in most marsupial mammals as well as many reptiles. Here we integrate data from the literature and first-hand studies of fossil and recent skeletal remains to reconstruct the evolution of the mammalian patella. We infer that bony patellae most likely evolved between four to six times in crown group Mammalia: in monotremes, in the extinct multituberculates, in one or more stem-mammal genera outside of therian or eutherian mammals, and up to three times in therian mammals. -

Uropsilus, Talpidae): Implications for Taxonomy and Conservation Tao Wan1,2†, Kai He1,3† and Xue-Long Jiang1*
Wan et al. BMC Evolutionary Biology 2013, 13:232 http://www.biomedcentral.com/1471-2148/13/232 RESEARCH ARTICLE Open Access Multilocus phylogeny and cryptic diversity in Asian shrew-like moles (Uropsilus, Talpidae): implications for taxonomy and conservation Tao Wan1,2†, Kai He1,3† and Xue-Long Jiang1* Abstract Background: The genus Uropsilus comprises a group of terrestrial, montane mammals endemic to the Hengduan and adjacent mountains. These animals are the most primitive living talpids. The taxonomy has been primarily based on cursory morphological comparisons and the evolutionary affinities are little known. To provide insight into the systematics of this group, we estimated the first multi-locus phylogeny and conducted species delimitation, including taxon sampling throughout their distribution range. Results: We obtained two mitochondrial genes (~1, 985 bp) and eight nuclear genes (~4, 345 bp) from 56 specimens. Ten distinct evolutionary lineages were recovered from the three recognized species, eight of which were recognized as species/putative species. Five of these putative species were found to be masquerading as the gracile shrew mole. The divergence time estimation results indicated that climate change since the last Miocene and the uplift of the Himalayas may have resulted in the diversification and speciation of Uropsilus. Conclusions: The cryptic diversity found in this study indicated that the number of species is strongly underestimated under the current taxonomy. Two synonyms of gracilis (atronates and nivatus) should be given full species status, and the taxonomic status of another three potential species should be evaluated using extensive taxon sampling, comprehensive morphological, and morphometric approaches. Consequently, the conservation status of Uropsilus spp. -

Constraints on the Timescale of Animal Evolutionary History
Palaeontologia Electronica palaeo-electronica.org Constraints on the timescale of animal evolutionary history Michael J. Benton, Philip C.J. Donoghue, Robert J. Asher, Matt Friedman, Thomas J. Near, and Jakob Vinther ABSTRACT Dating the tree of life is a core endeavor in evolutionary biology. Rates of evolution are fundamental to nearly every evolutionary model and process. Rates need dates. There is much debate on the most appropriate and reasonable ways in which to date the tree of life, and recent work has highlighted some confusions and complexities that can be avoided. Whether phylogenetic trees are dated after they have been estab- lished, or as part of the process of tree finding, practitioners need to know which cali- brations to use. We emphasize the importance of identifying crown (not stem) fossils, levels of confidence in their attribution to the crown, current chronostratigraphic preci- sion, the primacy of the host geological formation and asymmetric confidence intervals. Here we present calibrations for 88 key nodes across the phylogeny of animals, rang- ing from the root of Metazoa to the last common ancestor of Homo sapiens. Close attention to detail is constantly required: for example, the classic bird-mammal date (base of crown Amniota) has often been given as 310-315 Ma; the 2014 international time scale indicates a minimum age of 318 Ma. Michael J. Benton. School of Earth Sciences, University of Bristol, Bristol, BS8 1RJ, U.K. [email protected] Philip C.J. Donoghue. School of Earth Sciences, University of Bristol, Bristol, BS8 1RJ, U.K. [email protected] Robert J. -

Genetic and Morphologic Diversity of the Moles (Talpomorpha, Talpidae, Mogera) from the Continental Far East
Received: 15 August 2018 | Revised: 23 December 2018 | Accepted: 26 December 2018 DOI: 10.1111/jzs.12272 ORIGINAL ARTICLE Genetic and morphologic diversity of the moles (Talpomorpha, Talpidae, Mogera) from the continental Far East Elena Zemlemerova1,2 | Alexey Abramov3 | Alexey Kryukov4 | Vladimir Lebedev5 | Mi-Sook Min6 | Seo-Jin Lee6 | Anna Bannikova2 1A.N. Severtsov Institute of Ecology and Evolution, Russian Academy of Sciences, Abstract Moscow, Russia Taxonomy of the East Asian moles of the genus Mogera is still controversial. Based on 2 Lomonosov Moscow State University, the sequence data of 12 nuclear genes and one mitochondrial gene, we examine ge- Moscow, Russia netic variation in the Mogera wogura species complex and demonstrate that M. ro- 3Zoological Institute, Russian Academy of Sciences, Saint Petersburg, Russia busta, from the continental Far East, and M. wogura, from the Japanese Islands, are 4Federal Scientific Center of the East not conspecific. Our data do not support the existence of two or more species of Asia Terrestrial Biodiversity, Far Eastern Branch, Russian Academy of Sciences, Mogera in the Russian Far East. We suggest that the form “coreana” from the Korean Vladivostok, Russia Peninsula should be treated as a subspecies of M. robusta. Our morphological analy- 5 Zoological Museum, Lomonosov Moscow sis shows that M. r. coreana differs from typical M. robusta, from Primorye, primarily State University, Moscow, Russia in its smaller size. We show that there is strong morphological variability among con- 6Conservation Genome Resource Bank for Korean Wildlife (CGRB), Research Institute tinental moles, which may be associated with ecological and geographical factors. for Veterinary Science, College of Veterinary The time since the split between M. -

The Oldest Platypus and Its Bearing on Divergence Timing of the Platypus and Echidna Clades
The oldest platypus and its bearing on divergence timing of the platypus and echidna clades Timothy Rowe*†, Thomas H. Rich‡§, Patricia Vickers-Rich§, Mark Springer¶, and Michael O. Woodburneʈ *Jackson School of Geosciences, University of Texas, C1100, Austin, TX 78712; ‡Museum Victoria, PO Box 666, Melbourne, Victoria 3001, Australia; §School of Geosciences, PO Box 28E, Monash University, Victoria 3800, Australia; ¶Department of Biology, University of California, Riverside, CA 92521; and ʈDepartment of Geology, Museum of Northern Arizona, Flagstaff, AZ 86001 Edited by David B. Wake, University of California, Berkeley, CA, and approved October 31, 2007 (received for review July 7, 2007) Monotremes have left a poor fossil record, and paleontology has broadly affect our understanding of early mammalian history, been virtually mute during two decades of discussion about with special implications for molecular clock estimates of basal molecular clock estimates of the timing of divergence between the divergence times. platypus and echidna clades. We describe evidence from high- Monotremata today comprises five species that form two resolution x-ray computed tomography indicating that Teinolo- distinct clades (16). The echidna clade includes one short-beaked phos, an Early Cretaceous fossil from Australia’s Flat Rocks locality species (Tachyglossus aculeatus; Australia and surrounding is- (121–112.5 Ma), lies within the crown clade Monotremata, as a lands) and three long-beaked species (Zaglossus bruijni, Z. basal platypus. Strict molecular clock estimates of the divergence bartoni, and Z. attenboroughi, all from New Guinea). The between platypus and echidnas range from 17 to 80 Ma, but platypus clade includes only Ornithorhynchus anatinus (Austra- Teinolophos suggests that the two monotreme clades were al- lia, Tasmania). -

Influence of Evolutionary Allometry on Rates of Morphological Evolution and Disparity in Strictly Subterranean Moles (Talpinae, Talpidae, Lipotyphla, Mammalia)
J Mammal Evol DOI 10.1007/s10914-016-9370-9 ORIGINAL PAPER Influence of Evolutionary Allometry on Rates of Morphological Evolution and Disparity in strictly Subterranean Moles (Talpinae, Talpidae, Lipotyphla, Mammalia) G. Sansalone1,2,3 & P. Colangelo 2,4 & T. Kotsakis1,2 & A. Loy2,5 & R. Castiglia6 & A. A. Bannikova7 & E. D. Zemlemerova7 & P. Piras8,9 # Springer Science+Business Media New York 2017 Abstract The adaptation to a particular function could di- size variables. Evolutionary allometric trajectories exhibited rectly influence the morphological evolution of an anatomi- convergence of humeral shape between the two tribes, even cal structure as well as its rates. The humeral morphology of when controlling for phylogeny, though a significant differ- moles (subfamily Talpinae) is highly modified in response to ences in the evolutionary rates was found between the two intense burrowing and fully fossorial lifestyle. However, lit- tribes. Talpini, unlike Scalopini, seem to have reached a tle is known of the evolutionary pathways that marked its robust fossorial morphology early during their evolution, diversification in the two highly fossorial moles tribes and their shape disparity did not change, if it did not de- Talpini and Scalopini. We used two-dimensional landmark- crease, through time. Furthermore, the basal Geotrypus spp. based geometric morphometrics and comparative methods to clearly set apart from the other highly fossorial moles, understand which factors influenced the rates and patterns of exhibiting a significant acceleration of evolutionary shifts the morphological evolution of the humerus in 53 extant and toward higher degree of fossorial adaptation. Our observa- extinct species of the Talpini (22 extant plus 12 extinct) and tions support the hypothesis that the evolution of allometry Scalopini(sixextantplus13extinct)tribes,foratotalof623 may reflect a biological demand (in this case functional) that humeri. -
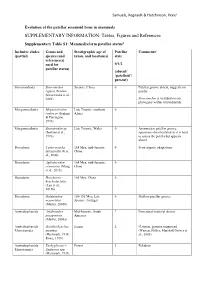
SUPPLEMENTARY INFORMATION: Tables, Figures and References
Samuels, Regnault & Hutchinson, PeerJ Evolution of the patellar sesamoid bone in mammals SUPPLEMENTARY INFORMATION: Tables, Figures and References Supplementary Table S1: Mammaliaform patellar status$ Inclusive clades Genus and Stratigraphic age of Patellar Comments# (partial) species (and taxon, and location(s) state reference(s) used for 0/1/2 patellar status) (absent/ ‘patelloid’/ present) Sinoconodonta Sinoconodon Jurassic, China 0 Patellar groove absent, suggests no rigneyi (Kielan- patella Jaworowska et al., 2004) Sinoconodon is included on our phylogeny within tritylodontids. Morganucodonta Megazostrodon Late Triassic, southern 0 rudnerae (Jenkins Africa & Parrington, 1976) Morganucodonta Eozostrodon sp. Late Triassic, Wales 0 Asymmetric patellar groove, (Jenkins et al., specimens disarticulated so it is hard 1976) to assess the patella but appears absent Docodonta Castorocauda 164 Mya, mid-Jurassic, 0 Semi-aquatic adaptations lutrasimilis (Ji et China al., 2006) Docodonta Agilodocodon 164 Mya, mid-Jurassic, 0 scansorius (Meng China et al., 2015) Docodonta Docofossor 160 Mya, China 0 brachydactylus (Luo et al., 2015b) Docodonta Haldanodon 150-155 Mya, Late 0 Shallow patellar groove exspectatus Jurassic, Portugal (Martin, 2005b) Australosphenida Asfaltomylos Mid-Jurassic, South ? Postcranial material absent patagonicus America (Martin, 2005a) Australosphenida Ornithorhynchus Extant 2 Platypus, genome sequenced Monotremata anatinus (Warren, Hillier, Marshall Graves et (Herzmark, 1938; al., 2008) Rowe, 1988) Australosphenida Tachyglossus -

Eutheria (Placental Mammals)
Eutheria (Placental Introductory article Mammals) Article Contents . Introduction J David Archibald, San Diego State University, San Diego, California, USA . Basic Design . Taxonomic and Ecological Diversity Eutheria includes one of three major clades of mammals, the extant members of which are . Fossil History and Distribution referred to as placentals. Phylogeny Introduction have supernumerary teeth (e.g. some whales, armadillos, Eutheria (or Placentalia) is the most taxonomically diverse etc.), in extant placentals the number of teeth is at most of three branches or clades of mammals, the other two three upper and lower incisors, one upper and lower being Metatheria (or Marsupialia) and Prototheria (or canine, four upper and lower premolars, and three upper Monotremata). When named by Gill in 1872, Eutheria and lower molars. Except for one fewer upper molar, a included both marsupials and placentals. It was Huxley in domestic dog retains this pattern. Compared to reptiles, 1880 that recognized Eutheria basically as used today to mammals have fewer skull bones through fusion and loss, include only placentals. McKenna and Bell in their although bones are variously emphasized in each of the Classification of Mammals, published in 1997, chose to three major mammalian taxa. use Placentalia rather than Eutheria to avoid the confusion Physiologically, mammals are all endotherms of varying of what taxa should be included in Eutheria. Others such as degrees of efficiency. They are also homeothermic with a Rougier have used Eutheria and Placentalia in the sense relatively high resting temperature. These characteristics used here. Placentalia includes all extant placentals and are also found in birds, but because of anatomical their most recent common ancestor. -

Author's Personal Copy
Author's personal copy Molecular Phylogenetics and Evolution 70 (2014) 513–521 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Multilocus phylogeny of talpine moles (Talpini, Talpidae, Eulipotyphla) and its implications for systematics ⇑ ⇑ Kai He a,b,1, Akio Shinohara c,1, Xue-Long Jiang a, , Kevin L. Campbell b, a State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650223, China b Department of Biological Sciences, University of Manitoba, Winnipeg, Manitoba R3T 2N2, Canada c Department of Bio-Resources, Division of Biotechnology, Frontier Science Research Center, University of Miyazaki, Miyazaki 889-1692, Japan article info abstract Article history: The tribe Talpini is a group of strictly subterranean moles distributed across the Eurasian Continent Received 5 June 2013 whose phylogenetic relationships and taxonomy remain unresolved. Here we report a multi-locus Revised 13 September 2013 nuclear-mitochondrial DNA dataset (9468 bp) from 11 talpine species encompassing all five recognized Accepted 3 October 2013 genera, together with analyses of their divergence times and evolutionary affinities inferred from maxi- Available online 16 October 2013 mum likelihood and Bayesian approaches. Our results finely resolved all relationships except the root of the four recognized Asian genera, which was placed sister to the genus Talpa. With respect to the Asian Keywords: clade, we moreover provide the first molecular support for a sister-taxon relationship between Parascap- Cryptic species tor and Scaptochirus and confirm that the genus Euroscaptor is paraphyletic. Further, and despite a rela- Species tree Species delimitation tively small sample size (22 specimens), our species delimitation analyses support the existence of at Talpidae least two genetically distinct, and hence potentially cryptic species. -

Phylogenetic Position of the Gansu Mole Scapanulus Oweni Thomas, 1912 and the Relationships Between Strictly Fossorial Tribes of the Family Talpidae1 A
ISSN 00124966, Doklady Biological Sciences, 2015, Vol. 464, pp. 230–234. © Pleiades Publishing, Ltd., 2015. Published in Russian in Doklady Akademii Nauk, 2015, Vol. 464, No. 2, pp. 238–242. GENERAL BIOLOGY Phylogenetic Position of the Gansu Mole Scapanulus oweni Thomas, 1912 and the Relationships between Strictly Fossorial Tribes of the Family Talpidae1 A. A. Bannikovaa, E. D. Zemlemerovaa, V. S. Lebedeva, D. Yu. Aleksandrovb, Yun Fangc, and B. I. Sheftelb Presented by Academician Yu. Yu. Dgebuadze April 6, 2015 Received April 9, 2015 Abstract—The results of the first molecular study focused on the phylogenetic position of the Gansu mole, Scapanulus oweni are presented. The analysis based on sequences of the mitochondrial cytb gene and five nuclear genes supports the monophyly of the Scalopini tribe including S. oweni and shows that two highly fos sorial talpid tribes, Talpini and Scalopini, are not immediate sister taxa. These results highlight the role of morphological parallelism as a potential source of conflict between molecular and morphologybased phy logenies in Talpidae. DOI: 10.1134/S0012496615050038 While essential progress was achieved during the could help to resolve phylogenetic relationships of this last years in the talpid molecular phylogenetics, many species. studies focus mainly on the relationships of species Concerning morphology, the available evidence is and genera within tribes [1–4]. The order of diver controversial in many respects. Myological data as well gence and relationships at the tribal level remains an as the parsimony analysis of the skull characters of the unsolved problem in the phylogenetic history of Talp presentday genera of Talpidae [7] do not support the idae. -

Condylura (Mammalia, Talpidae) Reloaded: New Insights About the Fossil Representatives of the Genus
Palaeontologia Electronica palaeo-electronica.org Condylura (Mammalia, Talpidae) reloaded: New insights about the fossil representatives of the genus Gabriele Sansalone, Tassos Kotsakis, and Paolo Piras ABSTRACT The star nosed mole, Condylura cristata, due to its morphological and behavioural peculiarities, has been deeply investigated by different authors. By contrast, very little is known about the phylogenetic relationships, evolution and diversity of the fossil members of this genus. In the present study we provide new insights about the fossil specimens ascribed to Condylura taking into account systematic, palaeobiogeographi- cal and palaeoecological aspects. Further, we provide a re-description of a fossil Con- dylura from the middle Miocene of Kazakhstan. We confirm that the Kazakh fossil belongs to the genus Condylura, based on humeral morphological features, and we discuss its implications and impact on the phylogenetic scenario and ecology of this peculiar talpid genus. This specimen represents the earliest record of the genus, thus suggesting an Eurasiatic origin instead of the most commonly accepted scenario of a North American one. The presence of both plesiomorphic and apomorphic characters in Condylura strongly supports the hypothesis that this genus could be considered as sister clade of Talpinae. Gabriele Sansalone. Roma Tre University of Rome, Dept. of Sciences, L.S. Murialdo, 1 – 00146 Rome, Italy/Center for evolutionary ecology, C.da Fonte Lappone, Pesche, Italy/Form, Evolution and Anatomy Research Laboratory, Zoology, School of Environmental and Rural Sciences, University of New England, Armidale, NSW 2351, Australia [email protected] Tassos Kotsakis. Roma Tre University of Rome, Dept. of Sciences, L.S. Murialdo, 1 – 00146 Rome, Italy/ Center for evolutionary ecology, C.da Fonte Lappone, Pesche, Italy [email protected] Paolo Piras. -

Talpid Mole Phylogeny Unites Shrew Moles and Illuminates Overlooked Cryptic Species Diversity Kai He,‡,†,1,2 Akio Shinohara,†,3 Kristofer M
Talpid Mole Phylogeny Unites Shrew Moles and Illuminates Overlooked Cryptic Species Diversity Kai He,‡,†,1,2 Akio Shinohara,†,3 Kristofer M. Helgen,4 Mark S. Springer,5 Xue-Long Jiang,*,1 and Kevin L. Campbell*,2 1State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming, China 2Department of Biological Sciences, University of Manitoba, Winnipeg, MN , Canada 3Department of Bio-resources, Division of Biotechnology, Frontier Science Research Center, University of Miyazaki, Miyazaki, Japan 4National Museum of Natural History Smithsonian Institution, Washington, DC 5Department of Biology, University of California, Riverside, CA ‡Present address: The Kyoto University Museum, Kyoto University, Kyoto, Japan †These authors contributed equally to this work. *Corresponding authors: E-mails: [email protected]; [email protected] Associate editor: Emma Teeling Abstract The mammalian family Talpidae (moles, shrew moles, desmans) is characterized by diverse ecomorphologies associated with terrestrial, semi-aquatic, semi-fossorial, fossorial, and aquatic-fossorial lifestyles. Prominent specializations involved with these different lifestyles, and the transitions between them, pose outstanding questions regarding the evolutionary history within the family, not only for living but also for fossil taxa. Here, we investigate the phylogenetic relationships, divergence times, and biogeographic history of the family using 19 nuclear and 2 mitochondrial genes (16 kb) from 60% of described species representing all 17 genera. Our phylogenetic analyses help settle classical questions in the evolution of moles, identify an ancient (mid-Miocene) split within the monotypic genus Scaptonyx, and indicate that talpid species richness may be nearly 30% higher than previously recognized. Our results also uniformly support the monophyly of long-tailed moles with the two shrew mole tribes and confirm that the Gansu mole is the sole living Asian member of an otherwise North American radiation.