Identification of Genes Associated With
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diffractive Properties of Blue Morpho Butterfly Wings
Diffractive Properties of Blue Morpho Butterfly Wings Mary Lalak and Paul Brackman Department of Physics and Astronomy, University of Georgia, Athens, Georgia 30602 (Dated: October 24, 2014) Many species of butterflies are known to produce beautiful iridescent colors when exposed to light from different angles. These affects can be attributed to a few different optical phenomena combined, the most prominent being reflective diffraction. Using common household items and with a budget of 50 dollars, we attempt to confirm the theory that the collection of ridges on the individual scales on the wing act as transmission gratings, as well as reflective. These results are quantified by the calculation of the ridge separation and comparing this distance to those observed by a scanning electron microscope photo. I. INTRODUCTION In studying the optical property of iridescence, three distinct mechanisms must be discussed. Thin film inter- ference, structured coloring, and reflective diffraction all contribute to the iridescent qualities of a surface. Thin film interference occurs when light strikes a film, some of it enters the film, and the rest is reflected off. The light transmitted through reflects off the bottom of the film and exits the film to interfere with the light that FIG. 1: Up-Close View of Scales from an Optical Microscope originally reflected off the surface of the film. This inter- ference pattern causes a spectrum of color to be visible from white light. Examples of thin film interference in- clude oil slicks and soap bubbles. Structural coloring oc- curs when the structure of the object itself (with various reflective surfaces) produces an interference resulting in vibrant colors. -

Iridescence in Cooked Venison – an Optical Phenomenon
Journal of Nutritional Health & Food Engineering Research Article Open Access Iridescence in cooked venison – an optical phenomenon Abstract Volume 8 Issue 2 - 2018 Iridescence in single myofibers from roast venison resembled multilayer interference in having multiple spectral peaks that were easily visible under water. The relationship HJ Swatland of iridescence to light scattering in roast venison was explored using the weighted- University of Guelph, Canada ordinate method of colorimetry. In iridescent myofibers, a reflectance ratio (400/700 nm) showing wavelength-dependent light scattering was correlated with HJ Swatland, Designation Professor CIE (Commission International de l’Éclairage) Y%, a measure of overall paleness Correspondence: Emeritus, University of Guelph, 33 Robinson Ave, Guelph, (r=0.48, P< 0.01). Hence, meat iridescence is an optical phenomenon. The underlying Ontario N1H 2Y8, Canada, Tel 519-821-7513, mechanism, subsurface multilayer interference, may be important for meat colorimetry. Email [email protected] venison, iridescence, interference, reflectance, meat color Keywords: Received: August 23, 2017 | Published: March 14, 2018 Introduction balanced pixel hues that have tricked your eyes to appear white; and when we perceive interference colors, complex interference spectra Iridescence is an enigmatic aspect of meat color with some practical trick our eyes again. As the order of interference increases, the colors 1–4 importance for consumers concerned about green colors in meat. appear to change from metallic -

Octopus Consciousness: the Role of Perceptual Richness
Review Octopus Consciousness: The Role of Perceptual Richness Jennifer Mather Department of Psychology, University of Lethbridge, Lethbridge, AB T1K 3M4, Canada; [email protected] Abstract: It is always difficult to even advance possible dimensions of consciousness, but Birch et al., 2020 have suggested four possible dimensions and this review discusses the first, perceptual richness, with relation to octopuses. They advance acuity, bandwidth, and categorization power as possible components. It is first necessary to realize that sensory richness does not automatically lead to perceptual richness and this capacity may not be accessed by consciousness. Octopuses do not discriminate light wavelength frequency (color) but rather its plane of polarization, a dimension that we do not understand. Their eyes are laterally placed on the head, leading to monocular vision and head movements that give a sequential rather than simultaneous view of items, possibly consciously planned. Details of control of the rich sensorimotor system of the arms, with 3/5 of the neurons of the nervous system, may normally not be accessed to the brain and thus to consciousness. The chromatophore-based skin appearance system is likely open loop, and not available to the octopus’ vision. Conversely, in a laboratory situation that is not ecologically valid for the octopus, learning about shapes and extents of visual figures was extensive and flexible, likely consciously planned. Similarly, octopuses’ local place in and navigation around space can be guided by light polarization plane and visual landmark location and is learned and monitored. The complex array of chemical cues delivered by water and on surfaces does not fit neatly into the components above and has barely been tested but might easily be described as perceptually rich. -
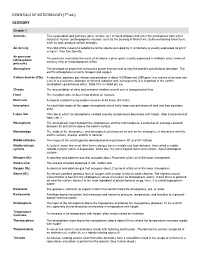
ESSENTIALS of METEOROLOGY (7Th Ed.) GLOSSARY
ESSENTIALS OF METEOROLOGY (7th ed.) GLOSSARY Chapter 1 Aerosols Tiny suspended solid particles (dust, smoke, etc.) or liquid droplets that enter the atmosphere from either natural or human (anthropogenic) sources, such as the burning of fossil fuels. Sulfur-containing fossil fuels, such as coal, produce sulfate aerosols. Air density The ratio of the mass of a substance to the volume occupied by it. Air density is usually expressed as g/cm3 or kg/m3. Also See Density. Air pressure The pressure exerted by the mass of air above a given point, usually expressed in millibars (mb), inches of (atmospheric mercury (Hg) or in hectopascals (hPa). pressure) Atmosphere The envelope of gases that surround a planet and are held to it by the planet's gravitational attraction. The earth's atmosphere is mainly nitrogen and oxygen. Carbon dioxide (CO2) A colorless, odorless gas whose concentration is about 0.039 percent (390 ppm) in a volume of air near sea level. It is a selective absorber of infrared radiation and, consequently, it is important in the earth's atmospheric greenhouse effect. Solid CO2 is called dry ice. Climate The accumulation of daily and seasonal weather events over a long period of time. Front The transition zone between two distinct air masses. Hurricane A tropical cyclone having winds in excess of 64 knots (74 mi/hr). Ionosphere An electrified region of the upper atmosphere where fairly large concentrations of ions and free electrons exist. Lapse rate The rate at which an atmospheric variable (usually temperature) decreases with height. (See Environmental lapse rate.) Mesosphere The atmospheric layer between the stratosphere and the thermosphere. -

Rainbow Peacock Spiders Inspire Miniature Super-Iridescent Optics
ARTICLE DOI: 10.1038/s41467-017-02451-x OPEN Rainbow peacock spiders inspire miniature super- iridescent optics Bor-Kai Hsiung 1,8, Radwanul Hasan Siddique 2, Doekele G. Stavenga 3, Jürgen C. Otto4, Michael C. Allen5, Ying Liu6, Yong-Feng Lu 6, Dimitri D. Deheyn 5, Matthew D. Shawkey 1,7 & Todd A. Blackledge1 Colour produced by wavelength-dependent light scattering is a key component of visual communication in nature and acts particularly strongly in visual signalling by structurally- 1234567890 coloured animals during courtship. Two miniature peacock spiders (Maratus robinsoni and M. chrysomelas) court females using tiny structured scales (~ 40 × 10 μm2) that reflect the full visual spectrum. Using TEM and optical modelling, we show that the spiders’ scales have 2D nanogratings on microscale 3D convex surfaces with at least twice the resolving power of a conventional 2D diffraction grating of the same period. Whereas the long optical path lengths required for light-dispersive components to resolve individual wavelengths constrain current spectrometers to bulky sizes, our nano-3D printed prototypes demonstrate that the design principle of the peacock spiders’ scales could inspire novel, miniature light-dispersive components. 1 Department of Biology and Integrated Bioscience Program, The University of Akron, Akron, OH 44325, USA. 2 Department of Medical Engineering, California Institute of Technology, Pasadena, CA 91125, USA. 3 Department of Computational Physics, University of Groningen, 9747 AG Groningen, The Netherlands. 4 19 Grevillea Avenue, St. Ives, NSW 2075, Australia. 5 Scripps Institution of Oceanography (SIO), University of California, San Diego, La Jolla, CA 92093, USA. 6 Department of Electrical and Computer Engineering, University of Nebraska-Lincoln, Lincoln, NE 68588, USA. -

Iridescent Color: from Nature to the Painter's Palette
Downloaded from http://www.mitpressjournals.org/doi/pdf/10.1162/LEON_a_00114 by guest on 25 September 2021 Technical ar T i c l e ce, Tech N e Iridescent Color: From Nature I , Sc to the Painter’s Palette T a b s T r a c T o: Ar nan The shifting rainbow hues of Franziska Schenk iridescence have, until recently, remained exclusive to nature. and Andrew Parker Now, the latest advances in nanotechnology enable the introduction of novel, bio- inspired color-shifting flakes into painting—thereby affording artists potential access to the full spectacle of iridescence. Unfortunately, existing rules rtists have never captured a color as dazzling selves of two distinct types: those of easel painting do not apply A that are stable attributes of material to the new medium; but, as and dynamic as the metallic blue of the Morpho butterfly, which nature inspired the technol- is visible for up to a quarter of a mile. Now, with rapid ad- substances, and those that are “acci- dental,” such as the evanescent col- ogy, an exploration of natural vances in nanoscience and technology, we are beginning to ors of the rainbow and the colors of phenomena can best inform unravel nature’s ingenious manipulation of the flow of light. some birds’ feathers, which change how to overcome this hurdle. Scientific research into natural nanoscale architectures, ca- according to the viewpoint of the Thus, by adopting a biomimetic pable of producing eye-catching optical effects, has led to the spectator [2]. approach, this paper outlines the optical principles underly- development of an ever-expanding range of comparable syn- ing iridescence and provides thetic structures. -

Diffraction Shading Models for Iridescent Surfaces
DIFFRACTION SHADING MODELS FOR IRIDESCENT SURFACES Emmanuel Agu Francis S.Hill Jr Department of Computer Science Department of Electrical and Computer Engineering, Worcester Polytechnic Institute, University of Massachusetts, Amherst, Worcester, MA 01609, USA MA 01002, USA [email protected] [email protected] Abstract to the phenomenon of iridescence. Common sources of iridescent colors include diffraction gratings, opals and Diffraction and interference are optical phenomena some liquid crystals, hummingbird winds and some which split light into its component wavelengths, hence snakeskins. Diffraction, also referred to as a wavefront producing a full spectrum of iridescent colors. This splitting phenomenon is distinguished from paper develops comput er graphics models for iridescent interference, an amplitude-splitting phenomenon [2]. colors produced by diffractive media. Diffraction gratings, certain animal skins and the crystal structure A common problem in optics is that of determining the of some precious stones are known to produce outgoing light intensity, wavelength and color, given diffraction. Several techniques can be employed to that light is incident on the diffraction surface at a derive solutions to the diffraction problem including: certain angle and intensity and is comprised of specified (1)Electromagnetic boundary value methods wavelengths. In particular, a closed-form expression (2)Applying the Huygens-Fresnel principle (3)Applying relating incoming and outgoing light permits diffraction the Kirchoff-Fresnel theorem (4)Fourier optics. surfaces to be elegantly modelled in computer graphics. Previous work in developing diffraction models for computer graphics has used boundary value methods and Fourier optics but no models using Huygens- Several techniques have been employed in the optics Fresnel principle have been published. -

The Structure and Optical Characters of Iridescent Glass
Proc. Indian Acad. Sci. A9 371-381 (1939) The structure and optical characters of iridescent glass SIR C V RAMAN and V S RAJAGOPALAN Received April 29, 1939 1. Introduction In the museums of Europe will be found exhibited specimens of ancient glassware excavated from archaeological sites and distinguished by a very beautiful iridescence. Nearly a century ago, Sir David Brewster (1840, 1859, 1860, 1864) examined and described specimens of such glass. According to him, the iridescence of glass which has suffered decomposition is due to its lamellar structure; in other words, the material is an ensemble of thin films optically distinct from each other. In consequence of this structure, it exhibits a large reflecting power, presenting vivid colours both by reflection and by transmission, these being complementary. Brewster noticed that only rarely do the laminae possess specularly reflecting surfaces; in many cases their surfaces are rough due to the presence of a large number of small cavities which may lie separate or run into each other; in other cases again, according to him, the films are covered with concentric stratified structures presenting between crossed nicols the pheno- menon of a black cross. Brewster's explanation of the iridescence of ancient glass appears to have been accepted without question by all subsequent observers. Considerable interest naturally attaches to the question why the glass acquires with time a laminar structure which in its normal state it does not possess. It is a matter of common knowledge that glass under prolonged exposure to wind and weather or to the action of suitable chemical reagents often acquires a superficial iridescence, and attempts have not been lacking to accelerate this process of attack and thus to reproduce experimentally some of the structures observed and described by Brewster in ancient glass. -

National Weather Service Glossary Page 1 of 254 03/15/08 05:23:27 PM National Weather Service Glossary
National Weather Service Glossary Page 1 of 254 03/15/08 05:23:27 PM National Weather Service Glossary Source:http://www.weather.gov/glossary/ Table of Contents National Weather Service Glossary............................................................................................................2 #.............................................................................................................................................................2 A............................................................................................................................................................3 B..........................................................................................................................................................19 C..........................................................................................................................................................31 D..........................................................................................................................................................51 E...........................................................................................................................................................63 F...........................................................................................................................................................72 G..........................................................................................................................................................86 -

A Practical Extension to Microfacet Theory for the Modeling of Varying Iridescence Laurent Belcour, Pascal Barla
A Practical Extension to Microfacet Theory for the Modeling of Varying Iridescence Laurent Belcour, Pascal Barla To cite this version: Laurent Belcour, Pascal Barla. A Practical Extension to Microfacet Theory for the Modeling of Varying Iridescence. ACM Transactions on Graphics, Association for Computing Machinery, 2017, 36 (4), pp.65. 10.1145/3072959.3073620. hal-01518344v2 HAL Id: hal-01518344 https://hal.archives-ouvertes.fr/hal-01518344v2 Submitted on 20 Jul 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. A Practical Extension to Microfacet Theory for the Modeling of Varying Iridescence LAURENT BELCOUR, Unity Technologies PASCAL BARLA, Inria Classical microfacets Iridescent microfacets Goniochromatic eects Fig. 1. Material appearance such as that of leather is usually reproduced with microfacet models in computer graphics. A more realistic result is achieved by adding a thin-film coating that produces iridescent colors [Akin 2014]. We replace the classic Fresnel reflectance term with a new Airy reflectance term that accounts for iridescence due to thin-film interference. Our main contribution consists in an analytical integration of the high-frequency spectral oscillations exhibited by Airy reflectance, which is essential for practical rendering in RGB. -

Cephalopod Guidelines
Reference Resources Caveats from AAALAC’s Council on Accreditation regarding this resource: Guidelines for the Care and Welfare of Cephalopods in Research– A consensus based on an initiative by CephRes, FELASA and the Boyd Group *This reference was adopted by the Council on Accreditation with the following clarification and exceptions: The AAALAC International Council on Accreditation has adopted the “Guidelines for the Care and Welfare of Cephalopods in Research- A consensus based on an initiative by CephRes, FELASA and the Boyd Group” as a Reference Resource with the following two clarifications and one exception: Clarification: The acceptance of these guidelines as a Reference Resource by AAALAC International pertains only to the technical information provided, and not the regulatory stipulations or legal implications (e.g., European Directive 2010/63/EU) presented in this article. AAALAC International considers the information regarding the humane care of cephalopods, including capture, transport, housing, handling, disease detection/ prevention/treatment, survival surgery, husbandry and euthanasia of these sentient and highly intelligent invertebrate marine animals to be appropriate to apply during site visits. Although there are no current regulations or guidelines requiring oversight of the use of invertebrate species in research, teaching or testing in many countries, adhering to the principles of the 3Rs, justifying their use for research, commitment of appropriate resources and institutional oversight (IACUC or equivalent oversight body) is recommended for research activities involving these species. Clarification: Page 13 (4.2, Monitoring water quality) suggests that seawater parameters should be monitored and recorded at least daily, and that recorded information concerning the parameters that are monitored should be stored for at least 5 years. -

Iridescence: More Than Meets The
The First Annual Frontiers in Life Sciences Conference IRIDESCENCE More than Meets the Eye February 6 - 9, 2008 Arizona State University - School Of Life Sciences Old Main, Carson Ballroom, Tempe Campus photo credit: Melissa Meadows TABLE OF CONTENTS Overview . 2-3 Acknowledgements . 4-5 Daily Events . 6-7 Fashion Show . 8-11 Guest Speaker Biographies . 12-15 Conference organizers . 16 Oral Presentation Abstracts . 16-35 Poster Abstracts . 35-43 Subject Index . 44-45 photo credit: Tomatito26 | Dreamstime Stock Photos CONFERENCE OVERVIEW 2 A unique, integrative 4–day conference on iridescent colors in nature, Iridescence: More than Meets the Eye is a graduate student proposed and organized conference supported by the Frontiers in Life Sciences program in Arizona State University’s School of Life Sciences . This conference intends to connect diverse groups of researchers to catalyze synthetic cross– disciplinary discussions regarding iridescent coloration in nature, identify new avenues of research, and explore the potential for these stunning natural phenomena to provide novel insights in fields as divergent as materials science, sexual selection and primary science education . We invite you to join us for this exciting event February 6 – 9, 2008 at Old Main, Carson Ballroom, Tempe Campus . Each day of the conference will be dedicated to a specific area of study . We have invited twelve main speakers from all over the world and from disciplines ranging from biology to nanotechnology . The day will begin with a series of plenary–style talks by invited speakers, followed by shorter talks by other participants as time allows . Afternoon break–out sessions centered on the day’s topic will give interested participants a chance to discuss and share ideas, leading to new collaborations and several publications .