Antibacterial Activity and Mode of Action of Lactoquinomycin a from Streptomyces Bacillaris
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Novel Β-Lactamase Inhibitors: a Therapeutic Hope Against the Scourge of Multidrug Resistance
REVIEW ARTICLE published: 24 December 2013 doi: 10.3389/fmicb.2013.00392 Novel β-lactamase inhibitors: a therapeutic hope against the scourge of multidrug resistance Richard R. Watkins1,2 *, Krisztina M. Papp-Wallace 3,4 , Sarah M. Drawz 5 and Robert A. Bonomo3,4,6,7 * 1 Department of Internal Medicine, Northeast Ohio Medical University, Rootstown, OH, USA 2 Division of Infectious Diseases, Akron General Medical Center, Akron, OH, USA 3 Research Service, Louis Stokes Cleveland Department of Veterans Affairs Medical Center, Cleveland, OH, USA 4 Department of Medicine, Case Western Reserve University, Cleveland, OH, USA 5 Department of Lab Medicine and Pathology, University of Minnesota, Minneapolis, MN, USA 6 Pharmacology, Case Western Reserve University, Cleveland, OH, USA 7 Molecular Biology and Microbiology, Case Western Reserve University, Cleveland, OH, USA Edited by: The increasing incidence and prevalence of multi-drug resistance (MDR) among contem- Marcelo Tolmasky, California State porary Gram-negative bacteria represents a significant threat to human health. Since University Fullerton, USA their discovery, β-lactam antibiotics have been a major component of the armamentarium Reviewed by: against these serious pathogens. Unfortunately, a wide range of β-lactamase enzymes have Marcelo Tolmasky, California State University Fullerton, USA emerged that are capable of inactivating these powerful drugs. In the past 30 years, a major Gabriel Gutkind, Universidad de advancement in the battle against microbes has been the development of β-lactamase Buenos Aires, Argentina inhibitors, which restore the efficacy of β-lactam antibiotics (e.g., ampicillin/sulbactam, *Correspondence: amoxicillin/clavulanate, ticarcillin/clavulanate, and piperacillin/tazobactam). Unfortunately, Richard R. Watkins, Division of many newly discovered β-lactamases are not inactivated by currently available inhibitors. -

Dissertation Submitted in Partial Fulfillment of the Regulations
IDENTIFICATION OF GENES CODING FOR CARBAPENEMASES IN CLINICAL ISOLATES OF CARBAPENEM RESISTANT KLEBSIELLA SPECIES IN A TERTIARY CARE HOSPITAL Dissertation submitted in Partial fulfillment of the Regulations required for the award of M.D. DEGREE In MICROBIOLOGY– BRANCH IV The Tamil Nadu DR. M.G.R. MEDICAL UNIVERSITY Chennai MAY 2020. UNIVERSITY REGISTRATION NO: 201714252 CERTIFICATE This is to certify that the enclosed work “Identification of Genes Coding for Carbapenemases in Clinical Isolates of Carbapenem Resistant Klebsiella Species in a Tertiary Care Hospital” submitted by Dr.S.Sharon Dorothy to The Tamilnadu Dr. MGR Medical University is based on bonafide cases studied and analysed by the candidate in the Department of Microbiology, Coimbatore Medical College Hospital during the period from July 2018 to June 2019 under the guidance and supervision of Dr. N.Mythily, MD., Professor & HOD, Department of Microbiology and the conclusion reached in this study are her own. Guide Dr. N.MYTHILY, MD., Professor & HOD, Department of Microbiology, Coimbatore Medical College, Coimbatore. Dr. B. ASOKAN. M.S., M.Ch., Dr., N.MYTHILY, MD., Dean, Professor & HOD, Coimbatore Medical College and Hospital, Department of Microbiology, Coimbatore – 14. Coimbatore Medical College, Coimbatore – 14. DECLARATION I, Dr.S.Sharon Dorothy solemnly declare that the dissertation entitled “Identification of Genes Coding for Carbapenemases in Clinical Isolates of Carbapenem Resistant Klebsiella Species in a Tertiary Care Hospital” was done by me at Coimbatore Medical College Hospital, during the period from July 2018 to June 2019 under the guidance and supervision of Dr. N. Mythily, M.D., Professor & HOD, Department of Microbiology, Coimbatore Medical College, Coimbatore. -

NIH Public Access Author Manuscript Expert Opin Investig Drugs
NIH Public Access Author Manuscript Expert Opin Investig Drugs. Author manuscript; available in PMC 2011 February 1. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Expert Opin Investig Drugs. 2010 February ; 19(2): 215±234. doi:10.1517/13543780903505092. Newer Antibacterial Drugs for a New Century Gina Devasahayam1, W. Michael Scheld, and Paul S. Hoffman* Department of Medicine, Division of Infectious Diseases and International Health, University of Virginia Health System, Charlottesville, VA 22908 Abstract Antibacterial drug discovery and development has slowed considerably in recent years with novel classes discovered decades ago and regulatory approvals tougher to get. This article describes newer classes of antibacterial drugs introduced or approved after year 2000, their mechanisms of action/ resistance, improved analogs, spectrum of activity and clinical trials. It also discusses new compounds in development with novel mechanisms of action as well as novel unexploited bacterial targets and strategies which may pave the way for combating drug resistance and emerging pathogens in the 21st century. Keywords antibacterial; drug discovery; drug resistance Infectious diseases are one of the leading causes of death worldwide, especially in low and middle income (LMIC) countries where second line antibacterial drugs against resistant bacteria are generally unavailable or unaffordable. In upper income countries (UIC), the emergence of multi-drug resistance in both community and hospital acquired infections has outpaced development and delivery of new drugs to the clinic. Most recently, the emergence of carbapenem resistance among Klebsiella sp. and related Gram negative bacteria illustrates the magnitude of the problem, as these multi-drug resistant infections are associated with high mortality rates and few treatment options 1. -

Performance Standards for Antimicrobial Susceptibility Testing
26th Edition M100S Performance Standards for Antimicrobial Susceptibility Testing This document provides updated tables for the Clinical and Laboratory Standards Institute antimicrobial susceptibility testing standards M02-A12, M07-A10, and M11-A8. An informational supplement for global application developed through the Clinical and Laboratory Standards Institute consensus process. Licensedto:Landspitali-NationalUniversityHospital-DivisionofDiagnosticMedicine Thisdocumentisprotectedbycopyright.CLSIorder#0,Downloadedon12/28/2015. Clinical and Laboratory Standards Institute Setting the standard for quality in medical laboratory testing around the world. The Clinical and Laboratory Standards Institute (CLSI) is a not-for-profit membership organization that brings together the varied perspectives and expertise of the worldwide laboratory community for the advancement of a common cause: to foster excellence in laboratory medicine by developing and implementing medical laboratory standards and guidelines that help laboratories fulfill their responsibilities with efficiency, effectiveness, and global applicability. Consensus Process Consensus—the substantial agreement by materially affected, competent, and interested parties—is core to the development of all CLSI documents. It does not always connote unanimous agreement, but does mean that the participants in the development of a consensus document have considered and resolved all relevant objections and accept the resulting agreement. Commenting on Documents CLSI documents undergo periodic evaluation and modification to keep pace with advancements in technologies, procedures, methods, and protocols affecting the laboratory or health care. CLSI’s consensus process depends on experts who volunteer to serve as contributing authors and/or as participants in the reviewing and commenting process. At the end of each comment period, the committee that developed the document is obligated to review all comments, respond in writing to all substantive comments, and revise the draft document as appropriate. -

Stembook 2018.Pdf
The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances FORMER DOCUMENT NUMBER: WHO/PHARM S/NOM 15 WHO/EMP/RHT/TSN/2018.1 © World Health Organization 2018 Some rights reserved. This work is available under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non-commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization. Suggested citation. The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances. Geneva: World Health Organization; 2018 (WHO/EMP/RHT/TSN/2018.1). Licence: CC BY-NC-SA 3.0 IGO. Cataloguing-in-Publication (CIP) data. -

New Quinone Antibiotics Against Methicillin-Resistant S. Aureus
antibiotics Article New Quinone Antibiotics against Methicillin-Resistant S. aureus Javier Campanini-Salinas 1,2,* , Juan Andrades-Lagos 1, Nicolás Hinojosa 1, Fabián Moreno 1, Pedro Alarcón 3, Gerardo González-Rocha 4,5 , Ian E. Burbulis 6,7 and David Vásquez-Velásquez 1,* 1 Drug Development Laboratory, Faculty of Chemical and Pharmaceutical, Sciences, Universidad de Chile, Sergio Livingstone 1007, Santiago 8380492, Chile; [email protected] (J.A.-L.); [email protected] (N.H.); [email protected] (F.M.) 2 Facultad de Medicina y Ciencia, Universidad San Sebastián, Lago Panguipulli 1390, Puerto Montt 5501842, Chile 3 Agents of Bacterial Meningitis Laboratory, Instituto de Salud Pública de Chile, Santiago 7780050, Chile; [email protected] 4 Laboratorio de Investigación en Agentes Antibacterianos (LIAA), Departamento de Microbiología, Facultad de Ciencias Biológicas, Universidad de Concepción, Concepción 4070386, Chile; [email protected] 5 Millennium Initiative for Collaborative Research on Bacterial Resistance (MICROB-R), Av. Las Condes 12.438, Lo Barnechea, Región Metropolitana, Santiago 8320000, Chile 6 Centro de Investigación Biomédica, Facultad de Medicina y Ciencias, Universidad San Sebastián, Sede de la Patagonia, Lago Panguipulli 1390, Puerto Montt 5501842, Chile; [email protected] 7 Department of Biochemistry and Molecular Genetics, University of Virginia School of Medicine, 1340 Jefferson Park Ave., Charlottesville, VA 22908, USA * Correspondence: [email protected] (J.C.-S.); [email protected] (D.V.-V.) Citation: Campanini-Salinas, J.; Abstract: There is an urgent need for the development of new antibiotics. Here, we describe the Andrades-Lagos, J.; Hinojosa, N.; inhibitory activity of new quinone compounds against methicillin-resistant Staphylococcus aureus ® ® Moreno, F.; Alarcón, P.; (ATCC 43300), methicillin-sensitive S. -

Development of Antibiotics for Gram-Negatives: Where Now?
Therapeutic Perspective Development of antibiotics for Gram-negatives: where now? Clin. Invest. (2011) 1(2), 211–227 The incidence of infections due to multidrug-resistance pathogens, such as Enterobacteriaceae, Pseudomonas aeruginosa or Acinetobacter spp. has been Matteo Bassetti†1, Francesca increasing. The paucity of antimicrobials active against multidrug- resistance Ginocchio1, Daniele Roberto strains are an important challenge. Novel anti-Gram-negative agents from Giacobbe1 & Malgorzata Mikulska1 old antimicrobial classes include b-lactamase inhibitors, cephalosporins, 1Division of Infectious Diseases, San Martino carbapenems, aminoglycosides, polymyxin analogs, tetracycline and mono- Hospital & University of Genoa, bactams. Among them, b-lactamase inhibitors seem the most promis- L.go R.Benzi 10, 16132 Genova, Italy † ing as they might restore the activity of already known b-lactams against Author for correspondence: Tel.: +39 010 555 5132 b-lactamase-producing strains. New classes of antimicrobials include bis- Fax: +39 010 353 7680 indoles, boron-containing antibacterial protein-synthesis inhibitors, outer E-mail: [email protected] membrane synthesis inhibitors, antimicrobial peptides and antibiotics targeting novel sites of the 50S ribosomal subunit. Although promising, they are still far from being introduced into clinical practice. Therefore, optimizing the use of current antibiotics and infection control policies are mandatory. Keywords: antimicrobial peptides • ESKAPE • extended-spectrum b-lactamases • metallo-b-lactamases • new b-lactamase inhibitors • new cephalosporins Multidrug resistance (MDR) is defined as nonsusceptibility to one or more antimicrobials in three or more antimicrobial classes, while strains nonsusceptible to all antimicrobials, including polymyxins and tigecycline, are classified as extreme drug-resistant strains [1]. MDR Gram-negative bacteria pose a serious and rapidly emerging threat to patients in healthcare settings and are especially common and problematic in some intensive care units [2]. -
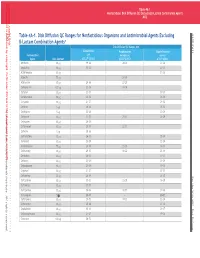
Table 4A-1. Disk Diffusion QC Ranges for Nonfastidious Organisms
This document is protected by copyright. CLSI order #Ord-615144, Downloaded on 1/13/2021. Downloaded on by CLSIorder#Ord-615144, document isprotected copyright. This Smit to:Juanita Licensed 7DEOH 1RQIDVLGLRVLVNLIIVLRQ4(FOGLQJβDFDPRPELQDLRQJHQV 0 Table 4A-1. Disk Diffusion QC Ranges for Nonfastidious Organisms and Antimicrobial Agents Excluding 0 β-Lactam Combination Agentsa Disk Diffusion QC Ranges, mm Escherichia Pseudomonas Staphylococcus Antimicrobial coli aeruginosa aureus Agent Disk Content ATCC®b 25922 ATCC® 27853 ATCC® 25923 Amikacin 30 mg 19–26 20–26 20–26 Ampicillin 10 mg 15–22 – 27–35 Azithromycin 15 mg – – 21–26 Azlocillin 75 mg – 24–30 – Aztreonam 30 mg 28–36 23–29 – Carbenicillin 100 mg 23–29 18–24 – Cefaclor 30 mg 23–27 – 27–31 Cefamandole 30 mg 26–32 – 26–34 Cefazolin 30 mg 21–27 – 29–35 Cefdinir m 24–28 – 25–32 5 g LLLL Cefditoren 5 mg 22–28 – 20–28 Cefepime 30 mg 31–37 25–31 23–29 Cefetamet 10 mg 24–29 – – Cefiderocol 30 mg 25–31 22–31 – Cefixime 5 mg 20–26 – – Cefmetazole 30 mg 26–32 – 25–34 Cefonicid 30 mg 25–29 – 22–28 Cefoperazone 75 mg 28–34 23–29 24–33 Cefotaxime 30 mg 29–35 18–22 25–31 Cefotetan 30 mg 28–34 – 17–23 Cefoxitin 30 mg 23–29 – 23–29 Cefpodoxime 10 mg 23–28 – 19–25 Cefprozil 30 mg 21–27 – 27–33 Ceftaroline 30 mg 26–34 – 26–35 UW0 Ceftazidime 30 mg 25–32 22–29 16–20 Ceftibuten 30 mg 27–35 – – Ceftizoxime 30 mg 30–36 12–17 27–35 Ceftobiprole 5 mg 25–31 – 20–27 Ceftriaxone 30 mg 29–35 17–23 22–28 Cefuroxime 30 mg 20–26 – 27–35 Cephalothin 30 mg 15–21 – 29–37 Chloramphenicol 30 mg 21–27 – 19–26 Cinoxacin 100 mg 26–32 – – This document is protected by copyright. -

Difficult to Treat Bacterial Infections: Have We Reached a “Dead End”?
Difficult to Treat Bacterial Infections: Have we reached a Dead End? Dr. George M. Varghese MD, DNB, DTMH, FRCP, FIDSA Department of Infectious Diseases Christian Medical College, Vellore, India Outline • Bacteria – well evolved for survival & lessens learned from history • Difficulty in treating due to inability in establishing the diagnosis • MDR gram negatives & treatment options • Will not discuss: – Difficult to eradiate infections eg. biofilm related – Immunologically protected sites – Immuno-compromised Bugs versus Drugs (and Humans) • The human body has 1013 human cells and 1014 bacteria! • Bacteria are the dominant species on the earth – Rapid multiplication rate – Natural mutation rate – Ability to transfer or move genes via plasmids, transposons etc. • Collectively, these properties allow bacteria to survive, change, and eventually flourish under intense selective pressure Search for a Magic bullet • Paul Ehrlich (1854 – 1915) • German bacteriologist • Father of Chemotherapy • Powerful weapon • Precise target • Selective destruction Discovery of Penicillin - 1928 • Penicillium notatum The Prophecy… • Dec 11, 1945: “There is the danger that the ignorant man may easily underdose himself and by exposing his microbes to non lethal quantities of the drug make them resistant” Antibiotic Resistance & Evolution PRP VRE VISA MRSA ESBL MBL VRSA 1961 1967 1983 1986 1988 1996 2002 Vancomycin Penicillin Vancomycin and teicoplanin 3rd gen Vancomycin β-lactams Carbapenem cephalosporin and teicoplanin Emergence → Spread Lechuguilla caves, New -

Disk Diffusion Susceptibility Test Procedure
HARDYDISK™ ANTIMICROBIAL SENSITIVITY TEST (AST) only INTENDED USE HardyDisk™ AST Disks are used for semi-quantitative in vitro susceptibility testing by the agar diffusion test procedure (Kirby- Bauer) of rapidly growing and certain fastidious bacterial pathogens. Standardized methods for agar diffusion testing have been described for Enterobacteriaceae, Staphylococcus spp., Pseudomonas spp., Acinetobacter spp., Listeria monocytogenes, Enterococcus spp., and by modified procedures, Haemophilus spp., Neisseria gonorrhoeae, N. meningitidis and Streptococcus spp., including Streptococcus pneumoniae.(5,6) SUMMARY Agar diffusion methods employing dried filter paper disks impregnated with specific concentrations of antimicrobial agents were developed in the 1940's. In order to eliminate or minimize variability in this testing, Bauer et al. developed a standardized procedure in which Mueller Hinton Agar was selected as the test medium.(1,2) Various regulatory agencies and standards-writing organizations subsequently published standardized reference procedures based on the Kirby-Bauer method. Among the earliest and most widely accepted of these standardized procedures were those published by the U.S. Food and Drug Administration (FDA) and the World Health Organization (WHO).(3-5) The procedure was adopted as a consensus standard by the Clinical and Laboratory Standards Institute (CLSI - formerly NCCLS) and is periodically updated.(6,7) Mueller Hinton Agar is currently recommended for disk diffusion testing of non-fastidious organisms such as Enterobacteriaceae, Staphylococcus spp., Pseudomonas spp., Acinetobacter spp., Listeria monocytogenes, Enterococcus spp. and other streptococci.(1,2) Using modified procedures, Haemophilus Test Medium (HTM) is now recommended for disk diffusion testing of Haemophilus species. Similarly, GC Base with Supplements is recommended for Neisseria gonorrhoeae and Mueller Hinton with 5% Sheep Blood is recommended for Streptococcus spp., and N. -

New Perspectives on Antibacterial Drug Research
Cent. Eur. J. Biol. • 8(10) • 2013 • 943-957 DOI: 10.2478/s11535-013-0209-6 Central European Journal of Biology New perspectives on antibacterial drug research Review Article Joanna Ziemska, Aleksandra Rajnisz, Jolanta Solecka* Laboratory of Biologically Active Compounds, National Institute of Public Health - National Institute of Hygiene, 00-791 Warsaw, Poland Received 14 March 2013; Accepted 10 May 2013 Abstract: Bacterial resistance to commonly used antibiotics is constantly increasing. Bacteria particularly dangerous for human life are methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus faecium and fluoroquinolone-resistant Pseudomonas aeruginosa. Hence, there is an incessant need for developing compounds with new modes of action and seeking alternate drug targets. In this review, the authors discuss the current situation of antibacterial medicines and present data on new antibiotic targets. Moreover, alternatives to antibiotics, such as bacteriophages, antimicrobial peptides and monoclonal antibodies, are presented. The authors also draw attention to the valuable features of natural sources in developing antibacterial compounds. The need to prevent and control infections as well as the reasonable use of currently available antibiotics is also emphasized. Keywords: Bacterial resistance • Antibacterial compound • Drug discovery • Target • Antimicrobial peptides © Versita Sp. z o.o. 1. Introduction pathogens (Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa) [2]. The use of antibiotics, especially the excessive and In this review, the authors will discuss the present indiscriminative use, both in medicine and veterinary status of bacterial resistance to antibiotics, especially science has contributed to the emergence of drug of bacterial species that cause serious hospital and resistant organisms. Antimicrobial drug resistance community-acquired infections. Furthermore, the constitutes a growing problem worldwide [1]. -

State of Antibiotic Development – Staying One Step Ahead?
Pfizer Animal Health State of Antibiotic Development – Staying one step ahead? Dr. Jeffrey L. Watts, PhD, RM (NRCM), M (ASCP) Director, Anti-Infectives Research 2012 One Medicine Symposium 5 – 6 December 2012 Durham, NC 1 Disclosures • Conflict of Interest Disclosure – Employee of Pfizer Animal Health – Pfizer Animal Health is engaged in the discovery, development and marketing of antibacterials used in companion animals and livestock 2 Presentation Topics • Overview of the Antibacterial Drug Discovery Processes • Antibacterial Drug Discovery in Human Health • Antibacterial Drug Discovery in Veterinary Medicine • The Search for Novel Substrate • Summary 3 The Antibacterial Discovery Process 4 Human and Animal Health R&D Processes Product Target Lead Candidate Preclinical Phase Phase Phase Approval Profile ID ID ID Data I II III First in Man Studies 12–15 years (Requires IND or Equivalent) 800M - 1.5 B Human Health Discovery First Target Animal Studies (Does not require INAD) Product Target Lead Candidate Preclinical Clinical Approval Profile ID ID ID Data Development 8–12 years 100 – 125 M Animal Health Discovery 5 Initiating the Discovery Process – Beginning with the end in mind Begins with a Target Profile • Label Claim (treatment of X disease caused by X organisms) • Market differentiators (single dose, oral, etc.) – Requires knowledge of current and future market conditions • Market value Key Points • Investment is made at risk • Assumes a defined regulatory process for that class of agent • Timeline for process is 10 – 15 for HH and