Protein Complex Formation in Methionine Chain-Elongation and Leucine Biosynthesis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Enzymatic Assay of L-METHIONINE GAMMA-LYASE (EC 4.4.1.11)
Enzymatic Assay of L-METHIONINE GAMMA-LYASE (EC 4.4.1.11) PRINCIPLE: L-Methionine Gamma-Lyase L-Methionine > Methanethiol + 2-Ketobutyrate + NH3 2-Ketobutyrate + MBTH > Azine Derivative Abbreviation used: MBTH = 3-Methyl-2-Benzothiazolinone CONDITIONS: T = 37°C, pH = 8.0, A320nm, Light path = 1 cm METHOD: Stopped Spectrophotometric Rate Determination REAGENTS: A. 100 mM Potassium Phosphate Buffer with 25 mM L-Methionine and 0.01 mM Pyridoxal 5-Phosphate, pH 8.0 at 37°C1 (Prepare 100 ml in deionized water using Potassium Phosphate, Monobasic, Anhydrous, Sigma Prod. No. P-5379, L-Methionine, Sigma Prod. No. M-9625, and Pyridoxal 5-Phosphate, Sigma Prod. No. P-9255. Adjust to pH 8.0 at 37°C with 5 M KOH.) B. 50% (w/v) Trichloroacetic Acid Solution (TCA) (Prepare 5 ml in deionized water using Trichloroacetic Acid, 6.1 N Solution, approximately 100% (w/v), Sigma Stock No. 490-10.) C. 1 M Sodium Acetate Buffer, pH 5.0 at 37°C (NaOAC) (Prepare 100 ml in deionized water using Sodium Acetate, Trihydrate, Sigma Prod. No. S-8625. Adjust to pH 5.0 at 37°C with 5 M HCl.) D. 0.1% (w/v) 3-Methyl-2-Benzothiazolinone Hydrazone (MBTH) (Prepare 10 ml in deionized water using 3-Methyl-2-Benzothiazolinone Hydrazone, Hydrochloride Hydrate, Sigma Prod. No. M-8006.) SSMETH01 Page 1 of 4 07/29/98 Enzymatic Assay of L-METHIONINE GAMMA-LYASE (EC 4.4.1.11) REAGENTS: E. 100 mM Potassium Phosphate Buffer with 1 mM Ethylenediaminetetraacetic Acid, 0.01% (v/v) 2-Mercaptoethanol and 0.02 mM Pyridoxal 5-Phosphate, pH 7.2 at 37°C (Enzyme Diluent)1 (Prepare 10 ml in deionized water using Potassium Phosphate, Monobasic, Anhydrous, Sigma Prod. -
![Lyxumia Subcutaneous Injection 300 Μg [Non-Proprietary Name] Lixisenatide (JAN*) [Name of Applicant] Sanofi K.K](https://docslib.b-cdn.net/cover/4198/lyxumia-subcutaneous-injection-300-g-non-proprietary-name-lixisenatide-jan-name-of-applicant-sanofi-k-k-324198.webp)
Lyxumia Subcutaneous Injection 300 Μg [Non-Proprietary Name] Lixisenatide (JAN*) [Name of Applicant] Sanofi K.K
Report on the Deliberation Results May 31, 2013 Evaluation and Licensing Division, Pharmaceutical and Food Safety Bureau Ministry of Health, Labour and Welfare [Brand name] Lyxumia Subcutaneous Injection 300 μg [Non-proprietary name] Lixisenatide (JAN*) [Name of applicant] Sanofi K.K. [Date of application] June 11, 2012 [Results of deliberation] In the meeting held on May 24, 2013, the First Committee on New Drugs concluded that the product may be approved and that this result should be reported to the Pharmaceutical Affairs Department of the Pharmaceutical Affairs and Food Sanitation Council. The re-examination period for the product is 8 years, and the drug substance and the drug product are both classified as powerful drugs and the product is not classified as a biological product or a specified biological product. The proposed Japanese brand name should be changed for ensuring medical safety. *Japanese Accepted Name (modified INN) This English version of the Japanese review report is intended to be a reference material to provide convenience for users. In the event of inconsistency between the Japanese original and this English translation, the former shall prevail. The PMDA will not be responsible for any consequence resulting from the use of this English version. Review Report May 7, 2013 Pharmaceuticals and Medical Devices Agency The results of a regulatory review conducted by the Pharmaceuticals and Medical Devices Agency on the following pharmaceutical product submitted for registration are as follows. [Brand name] Lyxumia Subcutaneous -

Effect of Leucine on Intestinal Absorption of Tryptophan in Rats
Downloaded from https://doi.org/10.1079/BJN19850155 British Journal of Nutrition (1985), 54, 695-703 695 https://www.cambridge.org/core Effect of leucine on intestinal absorption of tryptophan in rats BY CHISAE UMEZAWA, YUKO MAEDA, KANJI HABA, MARIKO SHIN AND KEIJI SANO School of Pharmacy, Kobe-Gakuin University, Nishi-ku, Kobe 673, Japan (Received I7 May 1985 - Accepted 24 June 1985) . IP address: 1. To elucidate the causal relation between leucine and the lowering of hepatic NAD content of rats fed on a leucine-excessive diet (Yamada et aZ. 1979), the effect of leucine on intestinal absorption of tryptophan was 170.106.35.93 investigated. 2. Co-administration of [3H]tryptophan and leucine, with leucine at ten times the level of tryptophan, delayed absorption of L-[side chain 2,3-3H]tryptophan from the digestive tract and incorporation of [3H]tryptophan into portal blood, the liver and a protein fraction of the liver. After 120 min, more than 95% of tryptophan was absorbed whether [3H]tryptophan was administered with or without leucine. , on 3. Co-administration of a mixture of ten essential amino acids, in proportions simulating casein, with 02 Oct 2021 at 04:49:27 [3H]tryptophan markedly delayed absorption of tryptophan from the digestive tract. The addition of supplementary leucine to the amino acid mixture, however, caused no further delay. 4. In rats prefed a leucine-excessive diet for 1 week [3H]tryptophan was absorbed at the same rate as in rats fed on a control diet. 5. The results indicate that competition between tryptophan and leucine for intestinal absorption did not cause lowering of hepatic NAD. -

Electrochemical Studies of Dl-Leucine, L-Proline and L
ELECTROCHEMICAL STUDIES OF DL -LEUCINE, 60 L-PROLINE AND L-TRYPTOPHAN AND THEIR INTERACTION WITH COPPER AND IRON 30 c b A) M. A. Jabbar, R. J. Mannan, S. Salauddin and B. µ a Rashid 0 Department of Chemistry, University of Dhaka, ( Current Dhaka-1000, Bangladesh -30 Introduction -60 In vitro study of the charge transfer reactions coupled -800 -400 0 400 800 with chemical reactions can give important indication of Potential vs. Ag/AgCl (mV) about actual biological processes occurring in human Fig.1. Comparison of the cyclic voltammogram of system. Understanding of such charge-transfer 5.0mM (a) DL -Leucine, (b) Cu-DL -Leucine ion and mechanism will help to determine the effectiveness of (c) [Fe-DL -Leucine] in 0.1M KCl solution at a Pt- nutrition, metabolism and treatment of various biological button electrode. Scan rates 50 mV/s. disorders. In the previous research, the redox behaviour of 40 various amino acids and biochemically important compounds and their charge transfer reaction and their b interaction of metal ions were studied [1,2]. In the present 20 ) a c research, the redox behavior and the charge transfer µΑ kinetics of DL -Leucine, L-Proline and L-Tryptophan in 0 the presence and absence of copper and iron will be investigated. Current ( -20 Experimental A computerized electrochemistry system developed by -40 -800 -400 0 400 800 Advanced Analytics, Virginia, USA, (Model-2040) Potential vs. Ag/AgCl (mV) consisting of three electrodes micro-cell with a saturated Ag/AgCl reference, a Pt-wire auxiliary and a pretreated Fig.2 . Comparison of the cyclic voltammogram of Pt-button working electrode is employed to investigate 5.0mM (a) L-Proline, (b) Cu-L-Proline and (c) Fe-L- Proline in 0.1M KCl solution at a Pt-button different amino acids and metal-amino acid systems. -

Amino Acid Chemistry
Handout 4 Amino Acid and Protein Chemistry ANSC 619 PHYSIOLOGICAL CHEMISTRY OF LIVESTOCK SPECIES Amino Acid Chemistry I. Chemistry of amino acids A. General amino acid structure + HN3- 1. All amino acids are carboxylic acids, i.e., they have a –COOH group at the #1 carbon. 2. All amino acids contain an amino group at the #2 carbon (may amino acids have a second amino group). 3. All amino acids are zwitterions – they contain both positive and negative charges at physiological pH. II. Essential and nonessential amino acids A. Nonessential amino acids: can make the carbon skeleton 1. From glycolysis. 2. From the TCA cycle. B. Nonessential if it can be made from an essential amino acid. 1. Amino acid "sparing". 2. May still be essential under some conditions. C. Essential amino acids 1. Branched chain amino acids (isoleucine, leucine and valine) 2. Lysine 3. Methionine 4. Phenyalanine 5. Threonine 6. Tryptophan 1 Handout 4 Amino Acid and Protein Chemistry D. Essential during rapid growth or for optimal health 1. Arginine 2. Histidine E. Nonessential amino acids 1. Alanine (from pyruvate) 2. Aspartate, asparagine (from oxaloacetate) 3. Cysteine (from serine and methionine) 4. Glutamate, glutamine (from α-ketoglutarate) 5. Glycine (from serine) 6. Proline (from glutamate) 7. Serine (from 3-phosphoglycerate) 8. Tyrosine (from phenylalanine) E. Nonessential and not required for protein synthesis 1. Hydroxyproline (made postranslationally from proline) 2. Hydroxylysine (made postranslationally from lysine) III. Acidic, basic, polar, and hydrophobic amino acids A. Acidic amino acids: amino acids that can donate a hydrogen ion (proton) and thereby decrease pH in an aqueous solution 1. -

The Un-Design and Design of Insulin: Structural Evolution
THE UN-DESIGN AND DESIGN OF INSULIN: STRUCTURAL EVOLUTION WITH APPLICATION TO THERAPEUTIC DESIGN By NISCHAY K. REGE Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Department of Biochemistry Dissertation Advisor: Dr. Michael A. Weiss CASE WESTERN RESERVE UNIVERSITY August, 2018 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of Nischay K. Rege candidate for the degree of Doctor of Philosophy*. Committee Chair Paul Carey Committee Member Michael Weiss Committee Member Faramarz Ismail-Beigi Committee Member George Dubyak Date of Defense: June 26th, 2018 *We also certify that written approval has been obtained for any proprietary material contained therein. Dedication This thesis is dedicated to my mother, Dipti, whose constant love and faith have never failed, to my father, Kiran, who taught me of the virtue of curiosity, and to my wife, Shipra, whose kindness and companionship have given me enough strength for eight lifetimes. i Table of Contents Dedication ..................................................................................................................................... i Table of Contents ......................................................................................................................... ii List of Tables ............................................................................................................................... v List of Figures ........................................................................................................................... -

Nucleotide Base Coding and Am1ino Acid Replacemients in Proteins* by Emil L
VOL. 48, 1962 BIOCHEMISTRY: E. L. SAIITH 677 18 Britten, R. J., and R. B. Roberts, Science, 131, 32 (1960). '9 Crestfield, A. M., K. C. Smith, and F. WV. Allen, J. Biol. Chem., 216, 185 (1955). 20 Gamow, G., Nature, 173, 318 (1954). 21 Brenner, S., these PROCEEDINGS, 43, 687 (1957). 22 Nirenberg, M. WV., J. H. Matthaei, and 0. WV. Jones, unpublished data. 23 Crick, F. H. C., L. Barnett, S. Brenner, and R. J. Watts-Tobin, Nature, 192, 1227 (1961). 24 Levene, P. A., and R. S. Tipson, J. Biol. Ch-nn., 111, 313 (1935). 25 Gierer, A., and K. W. Mundry, Nature, 182, 1437 (1958). 2' Tsugita, A., and H. Fraenkel-Conrat, J. Mllot. Biol., in press. 27 Tsugita, A., and H. Fraenkel-Conrat, personal communication. 28 Wittmann, H. G., Naturwissenschaften, 48, 729 (1961). 29 Freese, E., in Structure and Function of Genetic Elements, Brookhaven Symposia in Biology, no. 12 (1959), p. 63. NUCLEOTIDE BASE CODING AND AM1INO ACID REPLACEMIENTS IN PROTEINS* BY EMIL L. SMITHt LABORATORY FOR STUDY OF HEREDITARY AND METABOLIC DISORDERS AND THE DEPARTMENTS OF BIOLOGICAL CHEMISTRY AND MEDICINE, UNIVERSITY OF UTAH COLLEGE OF MEDICINE Communicated by Severo Ochoa, February 14, 1962 The problem of which bases of messenger or template RNA' specify the coding of amino acids in proteins has been largely elucidated by the use of synthetic polyri- bonucleotides.2-7 For these triplet nucleotide compositions (Table 1), it is of in- terest to examine some of the presently known cases of amino acid substitutions in polypeptides or proteins of known structure. -

Genetic Studies of Leptin Concentrations Implicate Leptin in the Regulation of Early
Page 1 of 93 Diabetes Genetic studies of leptin concentrations implicate leptin in the regulation of early adiposity Hanieh Yaghootkar1,2,3*&, Yiying Zhang4&, Cassandra N Spracklen5, Tugce Karaderi6,7,8,9, Lam Opal Huang10, Jonathan Bradfield11,12, Claudia Schurmann13, Rebecca S Fine14,15,16, Michael H Preuss13, Zoltan Kutalik1,17,18, Laura BL Wittemans6,19, Yingchang Lu13,20, Sophia Metz10, Sara M Willems19, Ruifang Li-Gao21, Niels Grarup10, Shuai Wang22, Sophie Molnos23,24, América A Sandoval-Zárate25, Mike A Nalls26,27, Leslie A Lange28, Jeffrey Haesser29, Xiuqing Guo30, Leo-Pekka Lyytikäinen31,32, Mary F Feitosa33, Colleen M Sitlani34, Cristina Venturini35, Anubha Mahajan6,36, Tim Kacprowski37,38, Carol A Wang22, Daniel I Chasman39,40, Najaf Amin41, Linda Broer42, Neil Robertson6,36, Kristin L Young43, Matthew Allison44, Paul L Auer45, Matthias Blüher46, Judith B Borja47,48, Jette Bork-Jensen10, Germán D Carrasquilla10, Paraskevi Christofidou35, Ayse Demirkan41, Claudia A Doege49, Melissa E Garcia50, Mariaelisa Graff43,51, Kaiying Guo4, Hakon Hakonarson11,52, Jaeyoung Hong22, Yii-Der Ida Chen30, Rebecca Jackson53, Hermina Jakupović10, Pekka Jousilahti54, Anne E Justice55, Mika Kähönen56,57, Jorge R Kizer58,59, Jennifer Kriebel23,24, Charles A LeDuc4, Jin Li60, Lars Lind61, Jian’an Luan19, David Mackey62, Massimo Mangino35,63, Satu Männistö54, Jayne F Martin Carli4, Carolina Medina-Gomez41,42, Dennis O Mook-Kanamori21,64, Andrew P Morris65,6,66, Renée de Mutsert21, Matthias Nauck67,38, Ivana Nedeljkovic41, Craig E Pennell22, Arund D Pradhan39,40, Bruce M Psaty68,69, Olli T Raitakari70,71,72, Robert A Scott19, Tea Skaaby73, Konstantin Strauch74,75, Kent D Taylor30, Alexander Teumer76,38, Andre G Uitterlinden41,42, Ying Wu5, Jie Yao30, Mark Walker77, Kari E North43, Peter Kovacs46, M. -

Amino Acid Transport Pathways in the Small Intestine of the Neonatal Rat
Pediat. Res. 6: 713-719 (1972) Amino acid neonate intestine transport, amino acid Amino Acid Transport Pathways in the Small Intestine of the Neonatal Rat J. F. FITZGERALD1431, S. REISER, AND P. A. CHRISTIANSEN Departments of Pediatrics, Medicine, and Biochemistry, and Gastrointestinal Research Laboratory, Indiana University School of Medicine and Veterans Administration Hospital, Indianapolis, Indiana, USA Extract The activity of amino acid transport pathways in the small intestine of the 2-day-old rat was investigated. Transport was determined by measuring the uptake of 1 mM con- centrations of various amino acids by intestinal segments after a 5- or 10-min incuba- tion and it was expressed as intracellular accumulation. The neutral amino acid transport pathway was well developed with intracellular accumulation values for leucine, isoleucine, valine, methionine, tryptophan, phenyl- alanine, tyrosine, and alanine ranging from 3.9-5.6 mM/5 min. The intracellular accumulation of the hydroxy-containing neutral amino acids threonine (essential) and serine (nonessential) were 2.7 mM/5 min, a value significantly lower than those of the other neutral amino acids. The accumulation of histidine was also well below the level for the other neutral amino acids (1.9 mM/5 min). The basic amino acid transport pathway was also operational with accumulation values for lysine, arginine and ornithine ranging from 1.7-2.0 mM/5 min. Accumulation of the essential amino acid lysine was not statistically different from that of nonessential ornithine. Ac- cumulation of aspartic and glutamic acid was only 0.24-0.28 mM/5 min indicating a very low activity of the acidic amino acid transport pathway. -
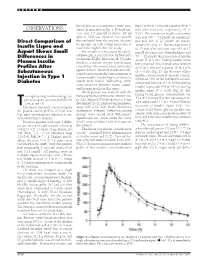
OBSERVATIONS Sured, As Indicated in Fig
LETTERS blood glucose concentrations were mea- lispro at 40 Ϯ 3 min and aspart at 49 Ϯ 3 OBSERVATIONS sured, as indicated in Fig. 1. If blood glu- min after injection, respectively (P ϭ cose was 3.5 mmol/l or lower, 20 ml 0.01). The maximum insulin concentra- glucose 30% was injected. One patient tion was 316 Ϯ 31 pmol/l on insulin lis- Direct Comparison of was excluded from the analysis because, pro and 295 Ϯ 27 pmol/l on insulin by mistake, he took a large extra dose of aspart (NS) (Fig. 1). The increase from 0 Insulin Lispro and insulin the night before the study. to 15 min after injection was 109 Ϯ 17 Aspart Shows Small Free insulin was measured after poly- pmol/l after injection of insulin lispro and Differences in ethylene glycol precipitation by Mercodia 53 Ϯ 11 pmol/l after injection of insulin Iso-Insulin (ELISA; Mercodia AB, Uppsala, Plasma Insulin aspart (P ϭ 0.02). Fasting insulin lispro Sweden), a two-site enzyme immunoassay levels reached 50% of peak concentration Profiles After containing two monoclonal antibodies at 20 Ϯ 1 min and aspart at 30 Ϯ 3 min against insulin. Identical results were ob- (P ϭ 0.02) (Fig. 2). The decrease of free Subcutaneous tained when equimolar concentrations of Injection in Type 1 insulin concentration from peak concen- human insulin, insulin lispro, and insulin tration to 50% of the maximum concen- aspart were tested, indicating 100% Diabetes tration was found at 113 Ϯ 10 min during cross-reactivity between lispro, aspart, insulin lispro and 154 Ϯ 14 min during and human insulin in this assay. -

Determination of Branched-Chain Keto Acids in Serum and Muscles Using High Performance Liquid Chromatography-Quadrupole Time-Of-Flight Mass Spectrometry
molecules Article Determination of Branched-Chain Keto Acids in Serum and Muscles Using High Performance Liquid Chromatography-Quadrupole Time-of-Flight Mass Spectrometry You Zhang, Bingjie Yin, Runxian Li and Pingli He * State Key Laboratory of Animal Nutrition, College of Animal Science and Technology, China Agricultural University, Beijing 100193, China; [email protected] (Y.Z.); [email protected] (B.Y.); [email protected] (R.L.) * Correspondence: [email protected]; Tel.: +86-10-6273-3688 Received: 4 December 2017; Accepted: 8 January 2018; Published: 11 January 2018 Abstract: Branched-chain keto acids (BCKAs) are derivatives from the first step in the metabolism of branched-chain amino acids (BCAAs) and can provide important information on animal health and disease. Here, a simple, reliable and effective method was developed for the determination of three BCKAs (α-ketoisocaproate, α-keto-β-methylvalerate and α-ketoisovalerate) in serum and muscle samples using high performance liquid chromatography-quadrupole time-of-flight mass spectrometry (HPLC-Q-TOF/MS). The samples were extracted using methanol and separated on a 1.8 µm Eclipse Plus C18 column within 10 min. The mobile phase was 10 mmol L−1 ammonium acetate aqueous solution and acetonitrile. The results showed that recoveries for the three BCKAs ranged from 78.4% to 114.3% with relative standard deviation (RSD) less than 9.7%. The limit of quantitation (LOQ) were 0.06~0.23 µmol L−1 and 0.09~0.27 nmol g−1 for serum and muscle samples, respectively. The proposed method can be applied to the determination of three BCKAs in animal serum and muscle samples. -

Utilization of Alpha-Ketoisocaproate for Protein Synthesis in Uremic Rats
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector Kidney International, Vol. 30 (1986), pp. 891—894 Utilization of alpha-ketoisocaproate for protein synthesis in uremic rats KRIANG TUNGSANGA, CHANG W. KANG, and MACKENZIE WALSER Department of Pharmacology and Molecular Science, and Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland U.S.A. Utilization of alpha-.ketoisocaproate for protein synthesis in uremic logues [1]. The reason for this was that studies of isolated rats. We have recently shown that the nutritional efficiency, R, ofperfused rat liver had shown that substantial fractions of each alpha—ketoisocaproate (KIC) as a substitute for leucine, defined as the ratio of the dose of leucine to the dose of KIC (on a leucine—free diet) BCKA were oxidized rather than transaminated [8]. for equal growth, can be evaluated isotopically: '4C-KIC and 3H-leucine Chawla, Stackhouse and Wadsworth [9] compared the are administered p.o.; six hours later, '4C/3H in the leucine of whole growth of rats fed aipha—ketoisocaproate (KIC) in place of body protein, divided by '4CPH in the injectate, gives a value indistin- leucine, and reported that four to five times as much KIC was guishable from R assessed in the same animals by growth experiments. To see how chronic uremia affects R, 11/12 nephrectomized rats and required to achieve equal growth. They defined nutritional sham—operated controls were fed a regular diet for 15 days and then efficiency of KIC as the ratio of the dose of leucine to the dose given these isotopes p.o.