A Protocol for Automated Timber Species Identification Using
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Physical, Mechanical, and Other Properties Of
ARC: 634.9 TA/OST 73-24 C559a PHYSICAL, MECHANICAL, AND OTHER PROPERTIES OF SELECTED SECONDARY SPECIES in Surinam, Peru, Colombia, Nigeria, Gabon, Philippines, and Malaysia FPL-AID-PASA TA(Aj)2-73 (Species Properties) * PHYSICAL, MECHANICAL, AND OTHER PROPERTIES OF SELECTED SECONDARY SPECIES LOCATED IN SURINAM, PERU, COLOMBIA, NIGERIA, GABON, PHILIPPINES, AND MALAYSIA MARTIN CHUDNOFF, Forest Products Technologist Forest Products Laboratory Forest Service, U.S. Department of Agriculture Madison, Wisconsin 53705 November 1973 Prepared for AGENCY FOR INTERNATIONAL DEVELOPMENT U.S. Department of State Washington, DC 20523 ARC No. 634.9 - C 559a INTRODUCTION This report is a partial response to a Participating Agency Service Agreement between the Agency for Inter national Development and the USDA, Forest Service (PASA Control No. TA(AJ)2-73) and concerns a study of the factors influencing the utilization of the tropical forest resource. The purpose of this portion of the PASA obligation is to present previously published information on the tree and wood characteristics of selected secondary species growing m seven tropical countries. The format is concise and follows the outline developed for the second edition of the "Handbook of Hardwoods" published by HMSO, London. Species selected for review are well known in the source countries, but make up a very small component, if any, of their export trade. The reasons why these species play a secondary role in the timber harvest are discussed in the other accompanying PASA reports. ii INDEX Pages SURINAM 1-11 Audira spp. Eperu falcata Eschweilera spp. Micropholis guyanensis Nectandra spp. Ocotea spp. Parinari campestris Parinari excelsa Pouteria engleri Protium spp. -

African Mahogany Anegre Birdseye Maple Black Walnut
African Mahogany African Mahogany (Khaya) is a genus of seven species of trees in the mahogany family Meliaceae, native to tropical Africa and Madagascar. All species become big trees 30-35 m tall, rarely 45 m, with a trunk over 1 m trunk diameter, often buttressed at the base. The leaves are pinnate, with 4-6 pairs of leaflets, the terminal leaflet absent; each leaflet is 10-15 cm long abruptly rounded toward the apex but often with an acuminate tip. The leaves can be either deciduous or evergreen depending on the species. The flowers are produced in loose inflorescences, each flower small, with four or five yellowish petals and ten stamens. The timber of Khaya is called African mahogany, the only timber widely accepted as mahogany besides that of the true mahogany, of the genus Swietenia. Khaya senegalensis, also known as the African dry zone mahogany is also used for its non timber parts. In West Africa, Fulani herdsmen prune the tree during the dry season to feed cattle. Anegre Anegre is milled from the Tawa tree (Beilschmiedia tawa) is a New Zealand broadleaf tree common in the central parts of the country. Tawa is often the dominant canopy species in lowland forests in the North Island and north east of the South Island, Individual specimens may grow up to 30 meters or more in height with trunks up to 1.2 meters in diameter, and they have smooth dark bark. The word "tawa" is the Maori name for the tree. One of the few hardwood trees in the country with good timber, the wood of this tree can be used for attractive and resilient floor boarding. -

Flexible Mating System in a Logged Population of Swietenia Macrophylla King (Meliaceae): Implications for the Management of a Threatened Neotropical Tree Species
Plant Ecol (2007) 192:169–179 DOI 10.1007/s11258-007-9322-9 ORIGINAL PAPER Flexible mating system in a logged population of Swietenia macrophylla King (Meliaceae): implications for the management of a threatened neotropical tree species Maristerra R. Lemes Æ Dario Grattapaglia Æ James Grogan Æ John Proctor Æ Roge´rio Gribel Received: 21 February 2007 / Accepted: 22 May 2007 / Published online: 19 June 2007 Ó Springer Science+Business Media B.V. 2007 Abstract Microsatellites were used to evaluate the crossed matings and that the remaining 6.75% had mating system of the remaining trees in a logged genotypes consistent with self-fertilisation. Apomixis population of Swietenia macrophylla, a highly valu- could be ruled out, since none of the 400 seedlings able and threatened hardwood species, in the Brazil- analysed had a multi-locus genotype identical to ian Amazon. A total of 25 open pollinated progeny its mother tree. The high estimate of the multi-locus arrays of 16 individuals, with their mother trees, were outcrossing rate (tm = 0.938 ± 0.009) using the mixed genotyped using eight highly polymorphic microsat- mating model also indicated that the population in ellite loci. Genotypic data analysis from the progeny this remnant stand of S. macrophylla was predomi- arrays showed that 373 out of the 400 seedlings nantly allogamous. The relatively large difference (93.25%) were unambiguously the result of out- between the multi-locus and single-locus outcrossing estimates (tmÀts = 0.117 ± 0.011) provides evidence that, in spite of the high outcrossing rate, a consid- & M. R. Lemes ( ) Á R. -

Complete Index of Common Names: Supplement to Tropical Timbers of the World (AH 607)
Complete Index of Common Names: Supplement to Tropical Timbers of the World (AH 607) by Nancy Ross Preface Since it was published in 1984, Tropical Timbers of the World has proven to be an extremely valuable reference to the properties and uses of tropical woods. It has been particularly valuable for the selection of species for specific products and as a reference for properties information that is important to effective pro- cessing and utilization of several hundred of the most commercially important tropical wood timbers. If a user of the book has only a common or trade name for a species and wishes to know its properties, the user must use the index of common names beginning on page 451. However, most tropical timbers have numerous common or trade names, depending upon the major region or local area of growth; furthermore, different species may be know by the same common name. Herein lies a minor weakness in Tropical Timbers of the World. The index generally contains only the one or two most frequently used common or trade names. If the common name known to the user is not one of those listed in the index, finding the species in the text is impossible other than by searching the book page by page. This process is too laborious to be practical because some species have 20 or more common names. This supplement provides a complete index of common or trade names. This index will prevent a user from erroneously concluding that the book does not contain a specific species because the common name known to the user does not happen to be in the existing index. -

Analysis on Trade Trend of CITES Regualted Tree Species and Its Impact on Chinese Smes
Analysis on Trade Trend of CITES Regualted Tree Species and its Impact on Chinese SMEs 2017.6 1 2 Preface The Convention on International Trade in Endangered Species of Wild Fauna and Flora (hereinafter referred to as CITES) is signed in Washington D.C. on 3 March 1973, also known as the Washington Convention. It entered into force on 1 July 1975. As an international convention on the control of international trade and the protection of wild flora and fauna, it aims to fully control the import and export of wild animals and plants as well as products thereof and manufactured goods through the regulation system, to promote protection and rational utilization of wild animals and plants resources to ensure that international trade in specimens of wild animals and plants does not threaten their survival. As of December 2016, CITES has 183 member countries where CITES legal framework and trade procedures are applied to regulate and monitor the international trade of species listed in CITES appendices. CITES has been in force for more than 40 years. It has been attracting more and more countries to jointly protect endangered wild animals and plants. At the same time, more and more fauna and flora species have been included in CITES appendices. Up to now, about 5,600 species of animals and 30,000 species of plants are protected by CITES against over-exploitation through international trade. In recent years, the sharp decline of endangered tree species has aroused widespread concern around the world, and over-exploitation is one of the main factors leading to this situation. -

The Woods of Liberia
THE WOODS OF LIBERIA October 1959 No. 2159 UNITED STATES DEPARTMENT OF AGRICULTURE FOREST PRODUCTS LABORATORY FOREST SERVICE MADISON 5, WISCONSIN In Cooperation with the University of Wisconsin THE WOODS OF LIBERIA1 By JEANNETTE M. KRYN, Botanist and E. W. FOBES, Forester Forest Products Laboratory,2 Forest Service U. S. Department of Agriculture - - - - Introduction The forests of Liberia represent a valuable resource to that country-- especially so because they are renewable. Under good management, these forests will continue to supply mankind with products long after mined resources are exhausted. The vast treeless areas elsewhere in Africa give added emphasis to the economic significance of the forests of Liberia and its neighboring countries in West Africa. The mature forests of Liberia are composed entirely of broadleaf or hardwood tree species. These forests probably covered more than 90 percent of the country in the past, but only about one-third is now covered with them. Another one-third is covered with young forests or reproduction referred to as low bush. The mature, or "high," forests are typical of tropical evergreen or rain forests where rainfall exceeds 60 inches per year without pro longed dry periods. Certain species of trees in these forests, such as the cotton tree, are deciduous even when growing in the coastal area of heaviest rainfall, which averages about 190 inches per year. Deciduous species become more prevalent as the rainfall decreases in the interior, where the driest areas average about 70 inches per year. 1The information here reported was prepared in cooperation with the International Cooperation Administration. 2 Maintained at Madison, Wis., in cooperation with the University of Wisconsin. -

Development of Nuclear SNP Markers for Mahogany (Swietenia Spp.)
Conservation Genetics Resources (2020) 12:585–587 https://doi.org/10.1007/s12686-020-01162-8 TECHNICAL NOTE Development of nuclear SNP markers for Mahogany (Swietenia spp.) Birte Pakull1 · Lasse Schindler1 · Malte Mader1 · Birgit Kersten1 · Celine Blanc‑Jolivet1 · Maike Paulini1 · Maristerra R. Lemes2 · Sheila E. Ward3 · Carlos M. Navarro4 · Stephen Cavers5,8 · Alexandre M. Sebbenn6 · Omar di Dio6 · Erwan Guichoux7 · Bernd Degen1 Received: 6 April 2020 / Accepted: 23 July 2020 / Published online: 12 August 2020 © The Author(s) 2020 Abstract Swietenia species are the most valuable American tropical timbers and have been heavily overexploited for decades. The three species are listed as either vulnerable or endangered by IUCN and are included on Appendix II of CITES, yet illegal exploitation continues. Here, we used restriction associated DNA sequencing to develop a new set of 120 SNP markers for Swietenia sp., suitable for MassARRAY®iPLEX™ genotyping. These markers can be used for population genetic studies and timber tracking purposes. Keywords SNPs · Mahogany · Swietenia spp. · MassARRAY®iPLEX™ The genus Swietenia includes the species: Swietenia mahag- commercially because of past overexploitation, S. macro- oni (L.) Jacq. (Small-leaved mahogany, native to Florida phylla is now the most valuable and economically important and the Caribbean islands), Swietenia macrophylla King. American tropical timber (Louppe et al. 2008). Swietenia (Big-leaved mahogany, native to Central and South Amer- wood is used for high-class furniture, boat building, musical ica) and Swietenia humilis Zucc. (Pacifc Coast mahogany, instruments etc. All three mahogany species are listed on native to the relatively dry Central American Pacifc coast) CITES (Convention on International Trade in Endangered (Schütt et al. -

African Mahogany Wood Defects Detected by Ultrasound Waves
General Technical Report FPL-GTR-239 • Proceedings: 19th International Nondestructive Testing and Evaluation of Wood Symposium African mahogany wood defects detected by ultrasound waves Tamara Suely Filgueira Amorim França Department of Sustainable Bioproducts, Mississippi State University, Starkville, Mississippi, United States, [email protected] Frederico Jose Nistal França Department of Sustainable Bioproducts, Mississippi State University, Starkville, Mississippi, United States, [email protected] Robert John Ross U.S. Forest Service, Forest Product Laboratory, Madison, Wisconsin, USA, [email protected] Xiping Wang U.S. Forest Service, Forest Product Laboratory, Madison, Wisconsin, USA, [email protected] Marina Donaria Chaves Arantes Departamento de Ciências Florestais e da Madeira, Universidade Federal do Espírito Santo, Jerônimo Monteira, Espírito Santo, Brasil, [email protected] Roy Daniel Seale Department of Sustainable Bioproducts, Mississippi State University, Starkville, Mississippi, United States, [email protected] Abstract This study aims to investigate the potential of ultrasound wave to detect defects in 19 years old of two species of African mahogany planted in Brazil. Were used five 76 x 5 x 5 cm samples from each species with different types of defects, and were conditioned to 12% moisture content. The samples were scanned with ultrasound wave in longitudinal direction and every 2,54 cm in radial and tangential directions along the samples. It was possible to identify end split and pin knots in Khaya ivorensis and reaction wood in Khaya senegalensis wood. Beetle galleries did not affect wave velocities in Khaya senegalensis wood. Grain angle had a large effect in ultrasound velocities in radial and tangential directions. Khaya senegalensis exhibit lower longitudinal velocities related to larger amount of interlocked grain in this species. -

Lumber / Veneer Match
Lumber Veneer Type of Cut Alder (Select) Alder Plain Sliced Face Alder (Knotty) Knotty Alder (Dark Putty in open knots) Plain Sliced Face Ash (Eastern White) Ash (Eastern White) Rotary Multi Piece Face Ash (Western) Ash (Eastern White) Rotary Multi Piece Face Beech (European Steamed) Beech (European Steamed) Plain Sliced Face Birch (Eastern Red) N/A N/A Birch (Eastern White) Birch (Eastern White) Rotary Multi Piece Face Birch (Western) N/A Cherry (Select) Cherry (Select) Plain Sliced Face Cherry (Rustic) Fir (CVG) Fir (CVG) Plain Sliced Face Hickory (Brown Heart) Hickory (Brown Heart) Plain Sliced Face Hickory (Calico) Hickory (Calico) Plain Sliced Face Hickory (Rustic Brown Heart) Hickory (Rustic Brown Heart) Plain Sliced Face Hickory (Rustic Calico) Hickory (Rustic Calico) Plain Sliced Face Lyptus Lyptus Plain Sliced Face Mahogany (African Khaya) Mahogany (African Khaya) Plain Sliced Face Mahogany (Edinam) Mahogany (African Khaya) Plain Sliced Face Maple (Eastern Hard White) Maple (Eastern Hard White) Rotary Whole Piece Face Maple (Eastern Soft Rustic) Maple (Eastern Hard Rustic) Plain Sliced Face Maple (Eastern Soft White) Maple (Eastern Hard White) Rotary Whole Piece Face Maple (Western) Alder Plain Sliced Face Paint Grade (Lumber Panel) DCD's Choice Rotary Whole Piece Face Paint Grade (MDF Panel) MDF N/A Pine (Eastern Clear) Pine (Eastern Clear) Plain Sliced Face Pine (Eastern Knotty) Pine (Eastern Knotty) Plain Sliced Face Poplar N/A N/A Red Oak (Natural) Red Oak (Natural) Plain Sliced Face Red Oak (Qtr Sawn) Red Oak (Qtr Sawn) Quarter Sawn Face Red Oak (Rift) Red Oak (Rift) Rift Sawn Face Red Oak (Select) Red Oak (Select) Plain Sliced Face Sapele (Flat Sawn) Sapele (Flat Sawn) Plain Sliced Face Sapele (Qtr Sawn) Sapele (Qtr Sawn) Quarter Sawn Face Walnut Walnut Plain Sliced Face White Oak (Select) White Oak (Select) Plain Sliced Face White Oak (Qtr Sawn) White Oak (Qtr Sawn) Quarter Sawn Face White Oak (Rift) White Oak (Rift) Rift Sawn Face. -

Luthier's Dictionary
LUTHIER’S DICTIONARY ©2014 The European Guitar Builders Association ©2014 The European Guitar Builders Association Woods ENGLISH DUTCH GERMAN FINNISH FRENCH ITALIAN SPANISH SWEDISH LATIN Alder Elzen Erle Leppä Aulne Ontano Aliso Al Alnus Alder / european, black, Elzen / Europees, Erle Leppä / eurooppalainen Aulne européen Ontano europeo Aliso Europeo Al (Klibbal) Alnus glutinosa common zwart, gewoon Alder / red, western red Elzen Erle Leppä / amerikkalainen Aulne rouge, Aulne Ontano americano Aliso Rojo Rödal Alnus rubra américain Arctic Birch Berken Birke Jääkoivu Bouleau arctique Betulla artica (bianca) Abedul del Ártico Björk Betula pendula Ash Essen Esche Saarni Frêne Frassino Fresno Ask Fraxinus Ash / american Amerikaans essen Esche Saarni / amerikkalainen Frêne américain Frassino americano Fresno Americano Vitask Fraxinus americana Ash / European Europees essen Esche Saarni / eurooppalainen Frêne européen Frassino europeo Fesno europeo Europeisk ask Fraxinus excelsior Ash / Swamp Ash Moeras essen Sumpfesche Suosaarni Frêne des marais Frassino palustre Fesno del Pantano Svartask Fraxinus nigra Basswood Linden Linde Lehmus Tilleul Tiglio Tilo Lind Tilia americana Bubinga Bubinga Bubinga Bubinga Bubinga Bubinga Bubinga Bubinga Guibourtia spp. (G. demeusei, G. pellegriniana, G. tessmannii, etc.) Cocobolo Cocobolo Cocobolo Cocobolo Coccobolo Cocobolo Cocobolo Mexikansk jakaranda Dalbergia retusa Ebony / African Afrikaans ebben Ebenholz Eebenpuu / Ébène Ebano africano Ébano Africano Ebenholts Diospyros crassiflora afrikkalainen Ebony / -

Population Genetic Structure of Mahogany (Swietenia Macrophylla
Molecular Ecology (2003) 12,2875-2883 doi: 10.1046/j.l365-294X.2003.01950.x Population genetic structure of mahogany (Swietenia macrophylla King, Meliaceae) across the Brazilian Amazon, based on variation at microsatellite loci: implications for conservation MARISTERRA R. LEMES,*t ROGERIO GRIBEL,*JOHN PROCTORt and DARIO GRATTAPAGLIAJ§ *Laboratorio de Genetica e Biologia Reprodutiva de Plantas, Institute) National de Pesquisas da Amazonia, C.P. 478, 69011-970 Manaus-AM, Brazil, ^Department of Biological Sciences, University of Stirling, Stirling FK9 4LA, UK, \haboraiorio de Genetica de Plantas, EMBRAPA-Recursos Geneticos e Biotecnologia, C.P. 02372, 70770-900 Brastlia-DF, Brazil, %Laboratorio de Biotecnologia Genomica, Pos-Graduaqao em Ciencias Genomicas, Universidade Catolica de Brasilia, SGAN 916 Mod. B, Asa Norte, 70790-160, Brasilia-DF, Brazil Abstract Mahogany (Swietenia macrophylla, Meliaceae) is the most valuable and intensively exploited Neotropical tree. No information is available regarding the genetic structure of mahogany in South America, yet the region harbours most of the unlogged populations of this prized hardwood. Here we report on the genetic diversity within and the differentiation among seven natural populations separated by up to 2100 km along the southern arc of the Brazilian Amazon basin. We analysed the variation at eight microsatellite loci for 194 adult individuals. All loci were highly variable, with the number of alleles per locus ranging from 13 to 27 (mean = 18.4). High levels of genetic diversity were found for all populations at the eight loci (mean ffE = 0.781, range 0.754-0.812). We found moderate but statistically significant genetic differentiation among populations considering both estimators of FST and RST, 9 = 0.097 and p = 0.147, respectively. -
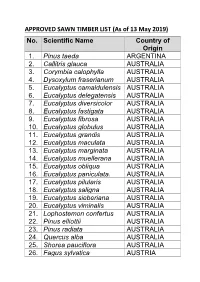
APPROVED SAWN TIMBER LIST (As of 13 May 2019) No. Scientific Name Country of Origin 1
APPROVED SAWN TIMBER LIST (As of 13 May 2019) No. Scientific Name Country of Origin 1. Pinus taeda ARGENTINA 2. Callitris glauca AUSTRALIA 3. Corymbia calophylla AUSTRALIA 4. Dysoxylum fraserianum AUSTRALIA 5. Eucalyptus camaldulensis AUSTRALIA 6. Eucalyptus delegatensis AUSTRALIA 7. Eucalyptus diversicolor AUSTRALIA 8. Eucalyptus fastigata AUSTRALIA 9. Eucalyptus fibrosa AUSTRALIA 10. Eucalyptus globulus AUSTRALIA 11. Eucalyptus grandis AUSTRALIA 12. Eucalyptus maculata AUSTRALIA 13. Eucalyptus marginata AUSTRALIA 14. Eucalyptus muellerana AUSTRALIA 15. Eucalyptus obliqua AUSTRALIA 16. Eucalyptus paniculata. AUSTRALIA 17. Eucalyptus pilularis AUSTRALIA 18. Eucalyptus saligna AUSTRALIA 19. Eucalyptus sieberiana AUSTRALIA 20. Eucalyptus viminalis AUSTRALIA 21. Lophostemon confertus AUSTRALIA 22. Pinus elliottii AUSTRALIA 23. Pinus radiata AUSTRALIA 24. Quercus alba AUSTRALIA 25. Shorea pauciflora AUSTRALIA 26. Fagus sylvatica AUSTRIA No. Scientific Name Country of Origin 27. Picea abies AUSTRIA 28. Picea abies BELARUS 29. Pinus sylvestris BELARUS 30. Quercus alba BELGIUM 31. Dipteryx odorata BOLIVIA 32. Apuleia leiocarpa BRAZIL 33. Astronium lecointei BRAZIL 34. Bagassa guianensis BRAZIL 35. Cedrela odorata BRAZIL 36. Cedrelinga catenaeformis BRAZIL 37. Couratari guianensis BRAZIL 38. Dipteryx odorata BRAZIL 39. Eucalyptus grandis BRAZIL 40. Eucalyptus grandis BRAZIL 41. Hymenaea courbaril BRAZIL 42. Hymenolobium modestum BRAZIL 43. Hymenolobium Nitidum BRAZIL Benth 44. Hymenolobium BRAZIL pulcherrimum 45. Manilkara bidentata BRAZIL 46. Myroxylon balsamum BRAZIL 47. Pinus radiata BRAZIL 48. Pinus taeda BRAZIL 49. Quassia simarouba BRAZIL 50. Tectona grandis BRAZIL 51. Fagus sylvatica BULGARIA No. Scientific Name Country of Origin 52. Quercus alba BULGARIA 53. Chlorophora excelsa CAMEROON 54. Cylicodiscus gabunensis CAMEROON 55. Entandrophragma CAMEROON candollei 56. Entandrophragma CAMEROON cylindricum 57. Entandrophragma CAMEROON cylindricum 58. Entandrophragma utile CAMEROON 59.