The Multifunctional Nucleolus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Size Changes in Eukaryotic Ribosomes
Proc. Nat. Acad. Sci. USA Vol. 68, No. 12, pp. 3021-3025, December 1971 Size Changes in Eukaryotic Ribosomes (diffusion constant/sedimentation constant/ribosomal dissociation/chick embryo) JOHN VOURNAKIS AND ALEXANDER RICH Department of Biology, Massachusetts Institute of Technology, Cambridge, Mass. 02139 Contributed by Alexander Rich, September 20, 1971 ABSTRACT Evidence is presented that ribosomes two particles are similar. However, these changes suggest active in protein synthesis and attached to messenger that when the ribosome is attached to messenger RNA it has a RNA on polysomes have a smaller diameter than free cytoplasmic single ribosomes. Measurements have been smaller diameter than is found for the free cytoplastic ribo- made on these two types of ribosomes of differences in some. This more compact form of the ribosome is maintained sedimentation velocity and diffusion constant. Differences even when the nascent polypeptide chain is relased by puro- in these quantities suggest about a 20-A decrease in the mycin. We thus infer that there are substantial differences diameter of the ribosomes from chick embryo muscles in the interactions between the ribosomal subunits when when they are attached to messenger RNA. Similar dif- they ferences are also observed in rabbit reticulocytes and are attached to messenger RNA as compared to the free cyto- mouse ascites tumor cells. These two ribosomal states plasmic single ribosome, which is inactive in protein synthesis. have different sensitivity to Pronase digestion and dis- sociate into ribosomal subunits at different KCI concen- METHODS AND MATERIALS trations. This size difference is not associated with a sig- nificant difference in overall ribosomal mass and appears Preparation of Ribosomes. -

A Global Analysis of Enzyme Compartmentalization to Glycosomes
pathogens Article A Global Analysis of Enzyme Compartmentalization to Glycosomes Hina Durrani 1, Marshall Hampton 2 , Jon N. Rumbley 3 and Sara L. Zimmer 1,* 1 Department of Biomedical Sciences, University of Minnesota Medical School, Duluth Campus, Duluth, MN 55812, USA; [email protected] 2 Mathematics & Statistics Department, University of Minnesota Duluth, Duluth, MN 55812, USA; [email protected] 3 College of Pharmacy, University of Minnesota, Duluth Campus, Duluth, MN 55812, USA; [email protected] * Correspondence: [email protected] Received: 25 March 2020; Accepted: 9 April 2020; Published: 12 April 2020 Abstract: In kinetoplastids, the first seven steps of glycolysis are compartmentalized into a glycosome along with parts of other metabolic pathways. This organelle shares a common ancestor with the better-understood eukaryotic peroxisome. Much of our understanding of the emergence, evolution, and maintenance of glycosomes is limited to explorations of the dixenous parasites, including the enzymatic contents of the organelle. Our objective was to determine the extent that we could leverage existing studies in model kinetoplastids to determine the composition of glycosomes in species lacking evidence of experimental localization. These include diverse monoxenous species and dixenous species with very different hosts. For many of these, genome or transcriptome sequences are available. Our approach initiated with a meta-analysis of existing studies to generate a subset of enzymes with highest evidence of glycosome localization. From this dataset we extracted the best possible glycosome signal peptide identification scheme for in silico identification of glycosomal proteins from any kinetoplastid species. Validation suggested that a high glycosome localization score from our algorithm would be indicative of a glycosomal protein. -

Cilia and Flagella: from Discovery to Disease Dylan J
Dartmouth Undergraduate Journal of Science Volume 20 Article 2 Number 1 Assembly 2017 Cilia and Flagella: From Discovery to Disease Dylan J. Cahill Dylan Cahill, [email protected] Follow this and additional works at: https://digitalcommons.dartmouth.edu/dujs Part of the Engineering Commons, Life Sciences Commons, Medicine and Health Sciences Commons, Physical Sciences and Mathematics Commons, and the Social and Behavioral Sciences Commons Recommended Citation Cahill, Dylan J. (2017) "Cilia and Flagella: From Discovery to Disease," Dartmouth Undergraduate Journal of Science: Vol. 20 : No. 1 , Article 2. Available at: https://digitalcommons.dartmouth.edu/dujs/vol20/iss1/2 This Research Article is brought to you for free and open access by the Student-led Journals and Magazines at Dartmouth Digital Commons. It has been accepted for inclusion in Dartmouth Undergraduate Journal of Science by an authorized editor of Dartmouth Digital Commons. For more information, please contact [email protected]. BIOLOGY Cilia and Flagella: FromCilia and Discovery Flagella: to Disease From Discovery to Disease BY DYLAN CAHILL ‘18 Introduction certain insect sperm fagella (3, 5, 6). A unique Figure 1: Chlamydomonas intracellular transport mechanism known as reinhardtii, a single-celled, bi- In 1674, peering through the lens of a crude flagellate green alga, viewed intrafagellar transport is responsible for the light microscope, Antoni van Leeuwenhoek with a scanning electron assembly and maintenance of these organelles Chlamydomonas observed individual living cells for the frst time microscope. is (3, 6). Cilia and fagella are primarily composed a model organism in flagellar in history (1). He noted long, thin appendages of the protein tubulin, which polymerizes into dynamics and motility studies. -

The Flagellum and Flagellar Pocket of Trypanosomatids
Molecular & Biochemical Parasitology 115 (2001) 1–17 www.parasitology-online.com. Reviews: Parasite cell Biology: 1 The flagellum and flagellar pocket of trypanosomatids Scott M. Landfear *, Marina Ignatushchenko Department of Molecular Microbiology and Immunology, Oregon Health Sciences Uni6ersity, Portland, OR 97201, USA Received 9 November 2000; received in revised form 26 January 2001; accepted 5 March 2001 Abstract The flagellum and flagellar pocket are distinctive organelles present among all of the trypanosomatid protozoa. Currently, recognized functions for these organelles include generation of motility for the flagellum and dedicated secretory and endocytic activities for the flagellar pocket. The flagellar and flagellar pocket membranes have long been recognized as morphologically separate domains that are component parts of the plasma membrane that surrounds the entire cell. The structural and functional specialization of these two membranes has now been underscored by the identification of multiple proteins that are targeted selectively to each of these domains, and non-membrane proteins have also been identified that are targeted to the internal lumina of these organelles. Investigations on the functions of these organelle-specific proteins should continue to shed light on the unique biological activities of the flagellum and flagellar pocket. In addition, work has begun on identifying signals or modifications of these proteins that direct their targeting to the correct subcellular location. Future endeavors should further refine our knowledge of targeting signals and begin to dissect the molecular machinery involved in transporting and retaining each polypeptide at its designated cellular address. © 2001 Elsevier Science B.V. All rights reserved. Keywords: Trypanosomatid protozoa; Flagellum; Flagellar Pocket; Organelle-specific proteins; Review 1. -

Driving Nucleolar Assembly
Downloaded from genesdev.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press PERSPECTIVE Driving nucleolar assembly Kathleen L. McCann,1 and Susan J. Baserga1,2,3,4 1Department of Genetics, 2Department of Molecular Biophysics and Biochemistry, 3Department of Therapeutic Radiology, Yale University School of Medicine, New Haven, Connecticut 06520, USA In this issue of Genes & Development,Grobandcol- breaks down (open mitosis), transcription of the ribosomal leagues (pp. 220–230) identify the minimal molecular re- RNA (rRNA) is inhibited, and the nucleolus disassem- quirements to assemble a fully functional nucleolus in bles. Upon completion of mitosis, rRNA transcription human cells and demonstrate the importance of the is reinitiated within the NOR, ribosome biogenesis fac- nucleolar transcription factor upstream binding factor tors are recruited, and the nucleolus is assembled. Only (UBF) as a mitotic bookmark at the ribosomal DNA NORs that are actively engaged in transcription can (rDNA). direct nucleolar assembly (Hernandez-Verdun 2011). Consequently, the nucleolus truly appears to be ‘‘an organelle formed by the act of building a ribosome’’ (Melese and Xue 1995). In all eukaryotes, the nucleolus is an essential, non- To begin to reveal the molecular mechanisms of nucle- membrane-bound organelle within the nucleus. Assem- olar formation, McStay’s laboratory (Mais et al. 2005; Grob bled around the ribosomal DNA (rDNA), the nucleolus et al. 2014) has applied synthetic biology. In an earlier is the site of ribosome biogenesis. In addition to its role report, they introduced 6.4 kb of DNA repeat sequences in making ribosomes, the nucleolus functions in other from the intergenic spacer of the Xenopus ribosomal gene important cellular processes, including stress sensing, into a noncanonical site in the human genome. -

Plastid in Human Parasites
SCIENTIFIC CORRESPONDENCE being otherwise homo Plastid in human geneous. The 35-kb genome-containing organ parasites elle identified here did not escape the attention of early Sm - The discovery in malarial and toxo electron microscopists who plasmodial parasites of genes normally - not expecting the pres occurring in the photosynthetic organelle ence of a plastid in a proto of plants and algae has prompted specula zoan parasite like tion that these protozoans might harbour Toxoplasma - ascribed to it a vestigial plastid1• The plastid-like para various names, including site genes occur on an extrachromosomal, 'Hohlzylinder' (hollow cylin maternally inherited2, 35-kilobase DNA der), 'Golgi adjunct' and circle with an architecture reminiscent of 'grof3e Tilkuole mit kriiftiger that of plastid genomes3•4• Although the Wandung' (large vacuole 35-kb genome is distinct from the 6-7-kb with stout surrounds) ( see linear mitochondrial genome3-6, it is not refs cited in ref. 9). Our pre known where in the parasite cells the plas liminary experiments with tid-like genome resides. Plasmodium falciparum, the To determine whether a plastid is pre causative agent of the most sent, we used high-resolution in situ lethal form of malaria, hybridization7 to localize transcripts of a identify an organelle (not plastid-like 16S ribosomal RNA gene shown) which appears sim from Toxoplasma gondii8, the causative ilar to the T. gondii plastid. agent of toxoplasmosis. Transcripts accu The number of surrounding mulate in a small, ovoid organelle located membranes in the P. anterior to the nucleus in the mid-region falciparum plastid, and its of the cell (a, b in the figure). -

Membrane Structure and Function
Chapter 7 Membrane Structure and Function Lecture Outline Overview: Life at the Edge • The plasma membrane separates the living cell from its surroundings. • This thin barrier, 8 nm thick, controls traffic into and out of the cell. • Like all biological membranes, the plasma membrane is selectively permeable, allowing some substances to cross more easily than others. • The formation of a membrane that encloses a solution different from the surrounding solution while still permitting the uptake of nutrients and the elimination of waste products was a key event in the evolution of life. • The ability of the cell to discriminate in its chemical exchanges with its environment is fundamental to life. • It is the plasma membrane and its component molecules that make this selectivity possible. Concept 7.1 Cellular membranes are fluid mosaics of lipids and proteins. • The main macromolecules in membranes are lipids and proteins, but carbohydrates are also important. • The most abundant lipids are phospholipids. • Phospholipids and most other membrane constituents are amphipathic molecules, which have both hydrophobic and hydrophilic regions. Membrane models have evolved to fit new data. • The arrangement of phospholipids and proteins in biological membranes is described by the fluid mosaic model. • In this model, the membrane is a fluid structure with a “mosaic” of various proteins embedded in or attached to a double layer (bilayer) of phospholipids. • Models of membranes were developed long before membranes were first seen with electron microscopes in the 1950s. • In 1915, membranes isolated from red blood cells were chemically analyzed and found to be composed of lipids and proteins. • In 1925, E. -

Cell Organelle Review You Need…
Cell Organelle Review You need…. Table cleared White Board Marker Eraser Mouth Closed! Word Bank • Vacuole • Cell Membrane • Cell Wall • Chloroplast • Mitochondrion • Cytoplasm • Nucleus Click to watch cell organelle video • http://www.youtube.com/watch?v=- zafJKbMPA8&feature=bf_prev&list=PLn6MYtn BEzPt5jgU8uP8lYmvuLg0JyPwn Description: surrounds the cytoplasm Organelle: Cell Membrane Structure or Function Control center Control center Nucleus Function Larger in plant cells Larger in plant cells Vacuole Structure Removes waste Removes waste Cell Membrane or Lysosome Function Chromosome are found here Chromosome are found here Nucleus Structure Converts glucose (food) into energy Converts glucose (food) into energy Mitochondrion Function Gel-like liquid that fills a cell Gel-like liquid that fills a cell Cytoplasm Structure Allows things to enter and exit the cell Allows things to enter and exit the cell Cell Membrane (Plasma Membrane) Function Provides support Provides support Cell Wall Function Storage site for water, nutrients and waste Storage site for water, nutrients and waste Vacuole Function Contains stacks of discs that house chlorophyll Contains stacks of discs that house chlorophyll Chloroplast Structure Double membrane with inner folds Double membrane with inner folds Mitochondrion Structure Site of photosynthesis (where plant food is made) Site of photosynthesis (where plant food is made) Chloroplast Function Rigid, green, surrounds plant cells Rigid, green, surrounds plant cells Cell Wall Structure Suspends organelles Suspends organelles Cytoplasm Function No nucleus No nucleus Prokaryotic Has a nucleus Has a nucleus Eukaryotic • Create a Venn Diagram using the word bank to differentiate between plant and animal cells. Word Bank Large Vacuole Small Vacuole Mitochondrion Plant Cell Both Nucleus Animal Cell Wall Cell Cytoplasm Chloroplast Cell Membrane Round Shape Square Shape • Create a Venn Diagram using the word bank to differentiate between plant and animal cells. -

The Cell Membrane Surrounds All Living Cells and Is the Most
The Cell membrane surrounds all living cells and is the most important organelle, there is also a similar plasma membrane that surrounds all the organelles except for the ribosome. The membrane controls how and what substances can move in and out of the cell/organelle The structure of the membrane is often referred to as the “Fluid Mosaic Model”; this is because of the way it is structured It is composed of phospholipids, proteins, and carbohydrates, which are arranged in a fluid mosaic structure. The phospholipids are arranged in a “bilayer”. With their hydrophilic (water attracting) phosphate heads facing outwards and their hydrophobic (water fearing) tails facing in towards the middle of the bilayer. The hydrophobic layer acts as a barrier to all but the smallest molecules and effectively isolating the two sides of the membranes. Some membranes contain phospholipids with different fatty acids, which affect the strength and flexibility. Animal cells also have cholesterol linking the fatty acids together and so stabilising and strengthening then membrane The proteins usually span from one side of the bilayer to the other. These are called integral proteins. But some sit on one side of the bilayer, these are called peripheral proteins. Proteins comprise approximately 50% of the mass of the membrane. The integral proteins (ones which span across the whole bilayer) are usually involved in the transporting of substances across the membrane. The proteins that are on the inside of the bilayer are often attached to the cytoskeleton and are involved in maintaining the cell’s shape. They may also be enzymes for catalysing reactions. -

Biogenesis of Glycosomes of Trypanosoma Brucei
Proc. Natl. Acad. Sci. USA Vol. 85, pp. 2598-2602, April 1988 Cell Biology Biogenesis of glycosomes of Trypanosoma brucei: An in vitro model of 3-phosphoglycerate kinase import (in vitro transcription of cloned DNA/in vitro translation/proteinase K digestion/signal sequence) HARRY F. DOVEY*, MARILYN PARSONSt*, AND CHING C. WANG*§ *Department of Pharmaceutical Chemistry, University of California, School of Pharmacy, San Francisco, CA 94143; tSeattle Biomedical Research Institute, 4 Nickerson Street, Seattle, WA 98109; and tDepartment of Pathobiology, University of Washington, Seattle, WA 98195 Communicated by Y. W. Kan, November 23, 1987 (receivedfor review August 3, 1987) ABSTRACT Glycosomes are intracellular, membrane- ined thus far, in vitro translation ofmRNA yields GPs with the bound microbody organelles of trypanosomes and leishmania. same molecular weights as the mature products inside the Nine glycolytic enzymes are the major protein components of glycosome (6), suggesting that the import may not involve the glycosomes of Trypanosoma brucei long-slender blood- proteolytic processing. Thus, this process appears similar to stream forms. Glycosomal proteins are believed to be synthe- the biogenesis of other microbodies, such as the peroxisomes sized in the cytoplasm and inserted across the glycosomal of yeast and mammals (7-9) and the glyoxysomes of plants membrane posttranslationally. We have developed an in vitro (10), and differs from the biogenesis of mitochondria and protein import assay for the study of glycosomal biogenesis in chloroplasts where posttranslational protein import generally T. brucei. All nine glycosomal glycolytic enzymes were detect- involves proteolytic cleavage of specific leader sequences able by immunoprecipitation and gel analysis of radiolabeled (11-13). -
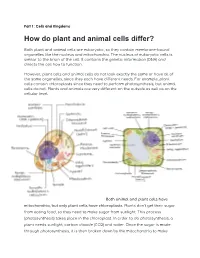
How Do Plant and Animal Cells Differ?
Part 1: Cells and Kingdoms How do plant and animal cells differ? Both plant and animal cells are eukaryotic, so they contain membrane-bound organelles like the nucleus and mitochondria. The nucleus of eukaryotic cells is similar to the brain of the cell. It contains the genetic information (DNA) and directs the cell how to function. However, plant cells and animal cells do not look exactly the same or have all of the same organelles, since they each have different needs. For example, plant cells contain chloroplasts since they need to perform photosynthesis, but animal cells do not. Plants and animals are very different on the outside as well as on the cellular level. Both animal and plant cells have mitochondria, but only plant cells have chloroplasts. Plants don’t get their sugar from eating food, so they need to make sugar from sunlight. This process (photosynthesis) takes place in the chloroplast. In order to do photosynthesis, a plant needs sunlight, carbon dioxide (CO2) and water. Once the sugar is made through photosynthesis, it is then broken down by the mitochondria to make energy for the cell. Because animals get sugar from the food they eat, they do not need chloroplasts: just mitochondria. Both plant and animal cells have vacuoles. A plant cell contains a large, singular vacuole that is used for storage of water and nutrients. It also helps maintain the shape of the cell. In contrast, animal cells have many, smaller vacuoles, which also are used for storage of water and nutrients. Plant cells have a cell wall, as well as a cell membrane. -

Cell Nucleus: the Nucleus
Available online at www.sciencedirect.com ScienceDirect Editorial overview: Cell nucleus: The nucleus: a dynamic organelle Michael P Rout and Gary H Karpen Current Opinion in Structural Biology 2014, 28:iv–vii For a complete overview see the Issue Available online 4th June 2014 http://dx.doi.org/10.1016/j.ceb.2014.05.005 0955-0674/Published by Elsevier Ltd. The nucleus is perhaps the defining feature of ‘eukaryotes’ (Greek ‘eu-’ Michael P Rout (with) ‘-karyon’ (kernel, nucleus)), and the emperor of all eukaryotic Laboratory of Cellular and Structural Biology, The Rockefeller University, New York, organelles in terms of scale and complexity of organization. It is, of course, NY 10065, United States the repository for almost all genomic information, encoded in DNA e-mail: [email protected] sequences wrapped into chromatin as discrete polymer packages termed chromosomes, surrounded in turn by a double-membrane nuclear envelope Michael P Rout received his Ph.D. in 1990 (NE). Our investment in genomics in humans and other eukaryotes has been from the Medical Research Council rightly huge; however, the return has been slow, as we discovered that Laboratory of Molecular Biology at the University of Cambridge, working with John sequence assemblies alone reveal little about how genetic information is Kilmartin. He joined Rockefeller in 1990 as a utilized. Instead, genome functions are greatly influenced by processes that postdoctoral fellow in Gu¨ nter Blobel’s control nuclear and chromosome architecture and dynamics — chromatin laboratory and was appointed assistant factors and modifications, epigenetic mechanisms, and pathways for con- professor and head of laboratory in 1997 and trolling transport of molecules into and out of the nucleus.