Bioinformatics and Evolution of Vertebrate Pancreatic Lipase And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rat CLPS / Colipase Protein (His Tag)
Rat CLPS / Colipase Protein (His Tag) Catalog Number: 80729-R08H General Information SDS-PAGE: Gene Name Synonym: CLPS Protein Construction: A DNA sequence encoding the rat Clps (NP_037271.1) (Met1-Gln112) was expressed with a polyhistidine tag at the C-terminus. Source: Rat Expression Host: HEK293 Cells QC Testing Purity: > 95 % as determined by SDS-PAGE. Endotoxin: Protein Description < 1.0 EU per μg protein as determined by the LAL method. Colipase belongs to the colipase family. Structural studies of the complex Stability: and of colipase alone have revealed the functionality of its architecture. It is a small protein with five conserved disulphide bonds. Structural analogies Samples are stable for up to twelve months from date of receipt at -70 ℃ have been recognised between a developmental protein, the pancreatic lipase C-terminal domain, the N-terminal domains of lipoxygenases and the Predicted N terminal: Ala 18 C-terminal domain of alpha-toxin. Colipase can only be detected in Molecular Mass: pancreatic acinar cells, suggesting regulation of expression by tissue- specific elements. Colipase allows lipase to anchor noncovalently to the The recombinant rat Clps consists 106 amino acids and predicts a surface of lipid micelles, counteracting the destabilizing influence of molecular mass of 11.9 kDa. intestinal bile salts. Without colipase the enzyme is washed off by bile salts, which have an inhibitory effect on the lipase. Colipase is a cofactor needed Formulation: by pancreatic lipase for efficient dietary lipid hydrolysis. It binds to the C- terminal, non-catalytic domain of lipase, thereby stabilising as active Lyophilized from sterile PBS, pH 7.4. -

Biochemical Properties of Pancreatic Colipase from the Common Stingray
Ben Bacha et al. Lipids in Health and Disease 2011, 10:69 http://www.lipidworld.com/content/10/1/69 RESEARCH Open Access Biochemical properties of pancreatic colipase from the common stingray Dasyatis pastinaca Abir Ben Bacha†, Aida Karray†, Lobna Daoud, Emna Bouchaala, Madiha Bou Ali, Youssef Gargouri and Yassine Ben Ali* Background: Pancreatic colipase is a required co-factor for pancreatic lipase, being necessary for its activity during hydrolysis of dietary triglycerides in the presence of bile salts. In the intestine, colipase is cleaved from a precursor molecule, procolipase, through the action of trypsin. This cleavage yields a peptide called enterostatin knoswn, being produced in equimolar proportions to colipase. Results: In this study, colipase from the common stingray Dasyatis pastinaca (CoSPL) was purified to homogeneity. The purified colipase is not glycosylated and has an apparent molecular mass of around 10 kDa. The NH2-terminal sequencing of purified CoSPL exhibits more than 55% identity with those of mammalian, bird or marine colipases. CoSPL was found to be less effective activator of bird and mammal pancreatic lipases than for the lipase from the same specie. The apparent dissociation constant (Kd) of the colipase/lipase complex and the apparent Vmax of the colipase-activated lipase values were deduced from the linear curves of the Scatchard plots. We concluded that Stingray Pancreatic Lipase (SPL) has higher ability to interact with colipase from the same species than with the mammal or bird ones. Conclusion: The fact that colipase is a universal lipase cofactor might thus be explained by a conservation of the colipase-lipase interaction site. -
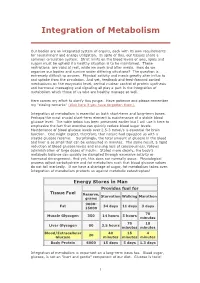
Integration of Metabolism
Integration of Metabolism Our bodies are an integrated system of organs, each with its own requirements for nourishment and energy utilization. In spite of this, our tissues share a common circulation system. Strict limits on the blood levels of ions, lipids and sugars must be upheld if a healthy situation is to be maintained. These restrictions are valid at rest, while we work and after meals. How do we organize our bodies and survive under differing situations? The question is extremely difficult to answer. Physical activity and meals greatly alter influx to and uptake from the circulation. And yet, feedback and feed-forward control mechanisms on the enzymatic level, central nuclear control of protein synthesis and hormonal messaging and signaling all play a part in the integration of metabolism which those of us who are healthy manage so well. Here comes my effort to clarify this jungle. Have patience and please remember my "closing remarks" (click here if you have forgotten them). Integration of metabolism is essential on both short-term and long-term bases. Perhaps the most crucial short-term element is maintenance of a stable blood glucose level. The table below has been presented earlier but I will use it here to emphasize the fact that exercise can quickly reduce blood sugar levels. Maintenance of blood glucose levels over 2.5-3 mmol/s is essential for brain function. One might expect, therefore, that nature had equipped us with a sizable glucose reserve. Surprisingly, the total amount of glucose in the blood and liver is so small that can be exhausted in minutes. -

Diagnostic Value of Serum Enzymes-A Review on Laboratory Investigations
Review Article ISSN 2250-0480 VOL 5/ ISSUE 4/OCT 2015 DIAGNOSTIC VALUE OF SERUM ENZYMES-A REVIEW ON LABORATORY INVESTIGATIONS. 1VIDYA SAGAR, M.SC., 2DR. VANDANA BERRY, MD AND DR.ROHIT J. CHAUDHARY, MD 1Vice Principal, Institute of Allied Health Sciences, Christian Medical College, Ludhiana 2Professor & Ex-Head of Microbiology Christian Medical College, Ludhiana 3Assistant Professor Department of Biochemistry Christian Medical College, Ludhiana ABSTRACT Enzymes are produced intracellularly, and released into the plasma and body fluids, where their activities can be measured by their abilities to accelerate the particular chemical reactions they catalyze. But different serum enzymes are raised when different tissues are damaged. So serum enzyme determination can be used both to detect cellular damage and to suggest its location in situ. Some of the biochemical markers such as alanine aminotransferase, aspartate aminotransferase, alkaline phasphatase, gamma glutamyl transferase, nucleotidase, ceruloplasmin, alpha fetoprotein, amylase, lipase, creatine phosphokinase and lactate dehydrogenase are mentioned to evaluate diseases of liver, pancreas, skeletal muscle, bone, etc. Such enzyme test may assist the physician in diagnosis and treatment. KEYWORDS: Liver Function tests, Serum Amylase, Lipase, CPK and LDH. INTRODUCTION mitochondrial AST is seen in extensive tissue necrosis during myocardial infarction and also in chronic Liver diseases like liver tissue degeneration DIAGNOSTIC SERUM ENZYME and necrosis². But lesser amounts are found in Enzymes are very helpful in the diagnosis of brain, pancreas and lung. Although GPT is plentiful cardiac, hepatic, pancreatic, muscular, skeltal and in the liver and occurs only in the small amount in malignant disorders. Serum for all enzyme tests the other tissues. -

Polymorphisms in PNLIP, Encoding Pancreatic Lipase, and Associations with Metabolic Traits
J Hum Genet (2001) 46:320–324 © Jpn Soc Hum Genet and Springer-Verlag 2001 ORIGINAL ARTICLE Robert A. Hegele · D. Dan Ramdath · Matthew R. Ban Michael N. Carruthers · Christine V. Carrington Henian Cao Polymorphisms in PNLIP, encoding pancreatic lipase, and associations with metabolic traits Received: January 18, 2001 / Accepted: February 19, 2001 Abstract Pancreatic lipase (EC 3.1.1.3) is an exocrine se- Introduction cretion that hydrolyzes dietary triglycerides in the small intestine. We developed genomic amplification primers to sequence the 13 exons of PNLIP, which encodes pancreatic Pancreatic lipase (PL; EC 3.1.1.3), a 56-kD protein, is in- lipase, in order to screen for possible mutations in cell lines volved in the intestinal hydrolysis of dietary triglyceride to of four children with pancreatic lipase deficiency (OMIM fatty acids. PL deficiency (OMIM 246600) is associated with 246600). We found no missense or nonsense mutations in malabsorption of long-chain triglyceride fatty acids, failure these samples, but we found three silent single-nucleotide to thrive in infancy and childhood, and absence of PL secre- polymorphisms (SNPs), namely, 96A/C in exon 3, 486C/T in tion upon secretin stimulation (Sheldon 1964; Rey et al. exon 6, and 1359C/T in exon 13. In 50 normolipidemic 1966). PNLIP on chromosome 10q26.1 (Davis et al. 1991) is Caucasians, the PNLIP 96C and 486T alleles had frequen- a 13-exon gene that spans more than 20kb (Sims et al. 1993) cies of 0.083 and 0.150, respectively. The PNLIP 1359T and encodes the 465-amino acid PL mature protein (Lowe allele was absent from Caucasian, Chinese, South Asian, et al. -

GC-MS Metabolic Profile and -Glucosidase-, -Amylase-, Lipase-, and Acetylcholinesterase-Inhibitory Activities of Eight Peach
molecules Article GC-MS Metabolic Profile and α-Glucosidase-, α-Amylase-, Lipase-, and Acetylcholinesterase-Inhibitory Activities of Eight Peach Varieties Dasha Mihaylova 1,* , Ivelina Desseva 2,* , Aneta Popova 3 , Ivayla Dincheva 4 , Radka Vrancheva 2 , Anna Lante 5 and Albert Krastanov 1 1 Department of Biotechnology, Technological Faculty, University of Food Technologies, 4002 Plovdiv, Bulgaria; [email protected] 2 Department of Analytical Chemistry and Physical Chemistry, Technological Faculty, University of Food Technologies, 4002 Plovdiv, Bulgaria; [email protected] 3 Department of Catering and Tourism, Economics Faculty, University of Food Technologies, 4002 Plovdiv, Bulgaria; [email protected] 4 AgroBioInstitute, Agricultural Academy, 8 Dr. Tsankov Blvd., 1164 Sofia, Bulgaria; [email protected] 5 Department of Agronomy, Food, Natural Resources, Animals, and Environment—DAFNAE, Agripolis, University of Padova, 35020 Legnaro, Italy; [email protected] * Correspondence: [email protected] (D.M.); [email protected] (I.D.) Abstract: The inhibition of certain digestive enzymes by target food matrices represents a new approach in the treatment of socially significant diseases. Proving the ability of fruits to inhibit such enzymes can support the inclusion of specific varieties in the daily diets of patients with diabetes, obesity, Alzheimer’s disease, etc., providing them with much more than just valuable Citation: Mihaylova, D.; Desseva, I.; micro- and macromolecules. The current study aimed atidentifying and comparing the GC-MS Popova, A.; Dincheva, I.; Vrancheva, metabolic profiles of eight peach varieties (“Filina”, “Ufo 4, “Gergana”, “Laskava”, “July Lady”, R.; Lante, A.; Krastanov, A. GC-MS “Flat Queen”, “Evmolpiya”, and “Morsiani 90”) grown in Bulgaria (local and introduced) and to Metabolic Profile and α-Glucosidase-, evaluate the inhibitory potential of their extracts towards α-glucosidase, α-amylase, lipase, and α-Amylase-, Lipase-, and acetylcholinesterase. -

The Role of Phospholipase D (Pld) and Grb2 in Chemotaxis
THE ROLE OF PHOSPHOLIPASE D (PLD) AND GRB2 IN CHEMOTAXIS A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science By KATIE J. KNAPEK B.S., Indiana University of Pennyslvania, 2006 2008 Wright State University WRIGHT STATE UNIVERSITY SCHOOL OF GRADUATE STUDIES December 19, 2008 I HEREBY RECOMMEND THAT THE THESIS PREPARED UNDER MY SUPERVISON BY Katie J. Knapek ENTITLED The Role of Phospholipase D (PLD) and Grb2 in Chemotaxis BE ACCEPTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science. _____________________________ Julian Gomez-Cambronero, Ph. D. Thesis Director _____________________________ Barbara Hull, Ph. D. Program Director Committee on Final Examination _______________________ Julian Gomez-Cambronero, Ph. D. _______________________ Nancy Bigley, Ph. D. _______________________ Mill Miller, Ph. D. ________________________ Joseph F. Thomas Jr., Ph. D. Dean, School of Graduate Studies ABSTRACT Knapek, Katie J. M.S., Department of Biological Sciences, Microbiology and Immunology Program, Wright State University, 2008. The Role of Phospholipase D (PLD) and Grb2 in Chemotaxis. Phospholipase D (PLD) is an enzyme that hydrolyzes phosphatidylcholine yielding choline and phosphatidic acid. PLD is activated by mitogens (lead to cell division) and motogens (leading to cell migration). PLD is known to contribute to cellular proliferation and deregulated expression of PLD has been implicated in several human cancers. PLD has been found to play a role in leukocyte chemotaxis and adhesion as studied through the formation of chemokine gradients. We have established a model of cell migration comprising three cell lines: macrophages RAW 264.7 and LR-5 (for innate defense), and fibroblast COS-7 cells (for wound healing). -

Multiple Roles of Phosphoinositide-Specific Phospholipase C Isozymes
BMB reports Mini Review Multiple roles of phosphoinositide-specific phospholipase C isozymes Pann-Ghill Suh1,*, Jae-Il Park1, Lucia Manzoli2, Lucio Cocco2, Joanna C. Peak3, Matilda Katan3, Kiyoko Fukami4, Tohru Kataoka5, Sanguk Yun1 & Sung Ho Ryu1 1Division of Molecular and Life Sciences, Pohang University of Science and Technology, Pohang, Korea, 2Cellular Signaling Laboratory, Department of Anatomical Sciences, University of Bologna, Via Irnerio, 48 I-40126, Bologna, Italy, 3Cancer Research UK Centre for Cell and Molecular Biology, Chester Beatty Laboratories, The Institute of Cancer Research, Fulham Road, London SW3 6JB, UK, 4Laboratory of Genome and Biosignal, Tokyo University of Pharmacy and Life Science, 1432-1 Horinouchi, Hachioji, 192-0392 Tokyo, Japan, 5Division of Molecular Biology, Department of Biochemistry and Molecular Biology, Kobe University Graduate School of Medicine, Chuo-ku, Kobe, Japan Phosphoinositide-specific phospholipase C is an effector mole- cellular calcium release (1-3; Fig. 1a). cule in the signal transduction process. It generates two sec- The first evidence of PLC activity was suggested by Hokin et ond messengers, inositol-1,4,5-trisphosphate and diacylglycer- al. in 1953 who reported specific hydrolysis of phospholipids ol from phosphatidylinositol 4,5-bisphosphate. Currently, thir- in pigeon’s pancreas slices after cholinergic stimulation (4). teen mammal PLC isozymes have been identified, and they are The authors showed that the enhanced turnover of phosphor- divided into six groups: PLC-β, -γ, -δ, -ε, -ζ and -η. Sequence ylinositol groups of phosphatidylinositol occurred in cells as a analysis studies demonstrated that each isozyme has more response to a variety of stimuli. In 1983, Streb et al. -

Pancreatic Amylase and Lipase Plasma Concentrations Are
Diabetes Care Volume 38, May 2015 e71 Pancreatic Amylase and Lipase Plasma David P. Sonne,1 Tina Vilsbøll,1 and Filip K. Knop1,2 Concentrations Are Unaffected by Increments in Endogenous GLP-1 Levels Following Liquid Meal Tests Diabetes Care 2015;38:e71–e72 | DOI: 10.2337/dc14-2751 Recent findings have suggested that amylase and lipase in plasma following patients with type 2 diabetes (P , incretin-based therapies promote pan- the ingestion of oral glucose and three 0.05), whereas lipase concentrations creatic inflammation and possibly cell isocaloric and isovolemic liquid mealsd were similar. Neither of the enzymes proliferation within the endocrine and all of which exerted normal endogenous increased following nutrient ingestion, exocrine pancreas (1). However, these GLP-1 secretion (4)din patients with suggesting that postprandial elevations studies have been met with substan- type 2 diabetes and matched control of endogenous GLP-1 (two- to three- tive criticism based on technical and subjects. fold) cannot trigger enzyme release methodical issues (2). Nevertheless, Detailed description of the experimen- from the human pancreas, at least not incretin-based therapies seem to result tal procedures and subjects was provided acutely. in small increases (within normal range) previously (4). In short, pancreas-specific These results suggest that the obser- in plasma concentrations of amylase amylase and lipase concentrations were vation of elevated plasma amylase and andlipaseinpatientsreceivingthese measured in plasma from 15 patients lipase -

Functional Role of Pentose Phosphate Pathway and Glutamine in Cancer Cell Metabolism Ibrahim Halil Polat
Functional role of pentose phosphate pathway and glutamine in cancer cell metabolism Ibrahim Halil Polat To cite this version: Ibrahim Halil Polat. Functional role of pentose phosphate pathway and glutamine in cancer cell metabolism. Human health and pathology. Universitat de Barcelona, 2016. English. NNT : 2016GREAS031. tel-01722703 HAL Id: tel-01722703 https://tel.archives-ouvertes.fr/tel-01722703 Submitted on 5 Mar 2018 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THÈSE Pour obtenir le grade de DOCTEUR DE LA COMMUNAUTÉ UNIVERSITÉ GRENOBLE ALPES préparée dans le cadre d’une cotutelle entre la Communauté Université Grenoble Alpes et Université de Barcelona Spécialité : BIS-Biotechnologie, instrumentation, signal et imagerie pour la biologie, la médecine et l’environnement Arrêté ministériel : 25 mai 2016 Présentée par « Ibrahim Halil Polat » Thèse dirigée par « Philippe Sabatier » codirigée par « Marta Cascante » préparée au sein des Laboratoires « TIMC-IMAG » et « BQI » dans les Écoles Doctorales « EDISCE » et « Ecole Doctorale de l’Université de Barcelone » Rôle fonctionnel des pentoses phosphates et glutamine dans le métabolisme des cellules cancéreuses Thèse soutenue publiquement le « 4 novembre 2016 », devant le jury composé de : M. Santiago Imperial Rodenas Professeur à l’Université de Barcelone (Président) Mme Loranne Agius Professeur à l’Université de Newcastle Upon Tyne (Rapporteur) M. -

Human CLPS / Colipase Protein (His Tag)
Human CLPS / Colipase Protein (His Tag) Catalog Number: 13631-H08B General Information SDS-PAGE: Gene Name Synonym: CLPS Protein Construction: A DNA sequence encoding the human CLPS (P04118) (Met 1-Gln 112) was fused with a polyhistidine tag at the C-terminus. Source: Human Expression Host: Baculovirus-Insect Cells QC Testing Purity: > 90 % as determined by SDS-PAGE Endotoxin: Protein Description < 1.0 EU per μg of the protein as determined by the LAL method Colipase belongs to the colipase family. Structural studies of the complex Stability: and of colipase alone have revealed the functionality of its architecture. It is a small protein with five conserved disulphide bonds. Structural analogies ℃ Samples are stable for up to twelve months from date of receipt at -70 have been recognised between a developmental protein, the pancreatic lipase C-terminal domain, the N-terminal domains of lipoxygenases and the Ala 18 Predicted N terminal: C-terminal domain of alpha-toxin. Colipase can only be detected in Molecular Mass: pancreatic acinar cells, suggesting regulation of expression by tissue- specific elements. Colipase allows lipase to anchor noncovalently to the The recombinant human CLPS consists of 105 amino acids and predicts a surface of lipid micelles, counteracting the destabilizing influence of molecular mass of 11.5 kDa. It migrates as an approximately 12 KDa band in intestinal bile salts. Without colipase the enzyme is washed off by bile salts, SDS-PAGE under reducing conditions. which have an inhibitory effect on the lipase. Colipase is a cofactor needed by pancreatic lipase for efficient dietary lipid hydrolysis. It binds to the C- Formulation: terminal, non-catalytic domain of lipase, thereby stabilising as active conformation and considerably increasing the overall hydrophobic binding Lyophilized from sterile PBS, 500mM NaCl, pH 7.0, 10% gly site. -

Unless Otherwise Noted, This Material Is Made Available
Author(s): Aken Desai, Michael Mathis, 2008 License: Unless otherwise noted, this material is made available under the terms of the Creative Commons Attribution – Share Alike 3.0 License: http://creativecommons.org/licenses/by-sa/3.0/ We have reviewed this material in accordance with U.S. Copyright Law and have tried to maximize your ability to use, share, and adapt it. Copyright holders of content included in this material should contact [email protected] with any quesCons, correcCons, or clarificaon regarding the use of content. For more informaon about how to cite these materials visit hHp://open.umich.edu/educaon/about/terms-of-use. Student works are presented as is and may be an interpretaon of faculty members’ lectures or assignments. These student works are not a product of faculty members. Faculty do not guarantee the accuracy of student work nor endorse them in any way. Any medical informaon in this material is intended to inform and educate and is not a tool for self-diagnosis or a replacement for medical evaluaon, advice, diagnosis or treatment by a healthcare professional. Please speak to your physician if you have quesCons about your medical condiCon. Viewer discreon is advised: Some medical content is graphic and may not be suitable for all viewers. Fatty Acids ctd. Tuesday, January 15, 2008 10:00 AM 14. What is the role of pentose pathway? Why is it important in fatty acid biosynthesis? What regulates the pentose pathway? How are pentoses formed without the oxidation of glucose‐6‐phosphate? a. Role is to provide NADPH and convert glucose into 5 carbon sugars for other pathways b.