US5266490.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Expression Cloning of a Human Dual-Specificity Phosphatase TOSHIO ISHIBASHI, DONALD P
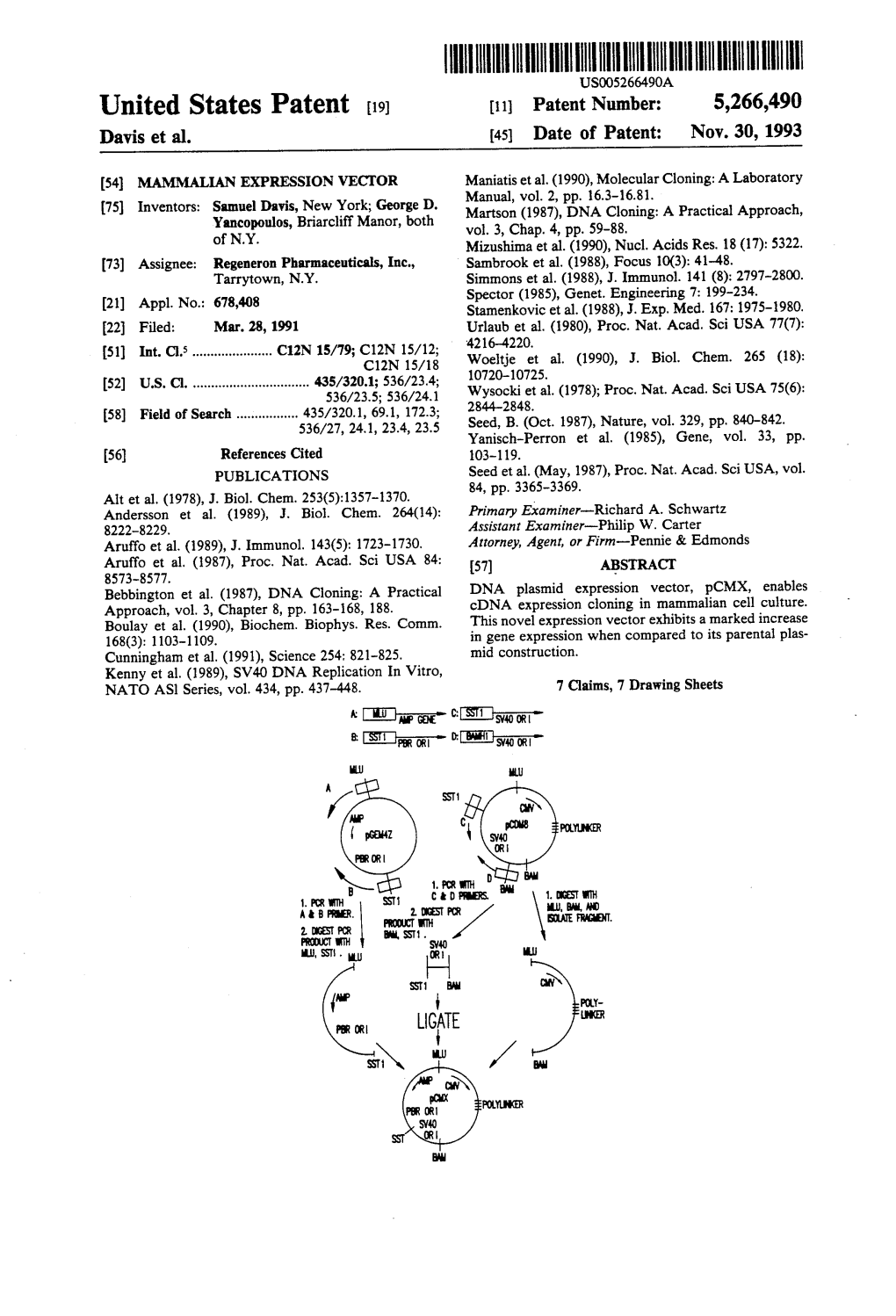
Proc. Nati. Acad. Sci. USA Vol. 89, pp. 12170-12174, December 1992 Biochemistry Expression cloning of a human dual-specificity phosphatase TOSHIO ISHIBASHI, DONALD P. BOTTARO, ANDREW CHAN, TORU MIKI, AND STUART A. AARONSON* Laboratory of Cellular and Molecular Biology, National Cancer Institute, Bethesda, MD 20892 Communicated by Leon A. Heppel, September 24, 1992 ABSTRACT Using an expression cloning strategy, we iso- substrate specificity for both phosphotyrosine and phospho- lated a cDNA encoding a human protein-tyrosine-phosphatase. serine. Bacteria expressing the kinase domain of the keratinocyte growth factor receptor (bek/fibroblast growth factor receptor 2) were infected with a fibroblast cDNA library in a phanid MATERIALS AND METHODS prokaryotic expression vector and screened with a monoclonal Plamid Construction of bek Kiame. The EcoRI-Sal I anti-phosphotyrosine antibody. Among several clones showing fragment of the mouse keratinocyte growth factor (KGF) decreased anti-phosphotyrosine recognition, one displayed receptor cDNA (15), which includes the kinase and carboxyl- phosphatase activity toward the kinase in vitro. The 4.1- terminal domains (residues 361-707), was ligated to the kilobase cDNA encoded a deduced protein of 185 amino ds EcoRP-Sal I fragment ofthe expression vector pCEV-lacZ to with limited sequence similar to the vaccinia virus phospha- generate the plasmid pCEV-bek (Fig. 1A). pCEV-lacZ is a tase VH1. The puriffied recombinant protein dephosphorlated pUC-based plasmid expression vector that produces a 3-gal several activated growth factor receptors, as well as serine- fusion protein under the control of the lac promoter (see phosphorylated casein, in vitro. Both serine and tyrosine phos- below). -

Temporal Proteomic Analysis of HIV Infection Reveals Remodelling of The
1 1 Temporal proteomic analysis of HIV infection reveals 2 remodelling of the host phosphoproteome 3 by lentiviral Vif variants 4 5 Edward JD Greenwood 1,2,*, Nicholas J Matheson1,2,*, Kim Wals1, Dick JH van den Boomen1, 6 Robin Antrobus1, James C Williamson1, Paul J Lehner1,* 7 1. Cambridge Institute for Medical Research, Department of Medicine, University of 8 Cambridge, Cambridge, CB2 0XY, UK. 9 2. These authors contributed equally to this work. 10 *Correspondence: [email protected]; [email protected]; [email protected] 11 12 Abstract 13 Viruses manipulate host factors to enhance their replication and evade cellular restriction. 14 We used multiplex tandem mass tag (TMT)-based whole cell proteomics to perform a 15 comprehensive time course analysis of >6,500 viral and cellular proteins during HIV 16 infection. To enable specific functional predictions, we categorized cellular proteins regulated 17 by HIV according to their patterns of temporal expression. We focussed on proteins depleted 18 with similar kinetics to APOBEC3C, and found the viral accessory protein Vif to be 19 necessary and sufficient for CUL5-dependent proteasomal degradation of all members of the 20 B56 family of regulatory subunits of the key cellular phosphatase PP2A (PPP2R5A-E). 21 Quantitative phosphoproteomic analysis of HIV-infected cells confirmed Vif-dependent 22 hyperphosphorylation of >200 cellular proteins, particularly substrates of the aurora kinases. 23 The ability of Vif to target PPP2R5 subunits is found in primate and non-primate lentiviral 2 24 lineages, and remodeling of the cellular phosphoproteome is therefore a second ancient and 25 conserved Vif function. -

Human Intestinal Nutrient Transporters
Gastrointestinal Functions, edited by Edgard E. Delvin and Michael J. Lentze. Nestle Nutrition Workshop Series. Pediatric Program. Vol. 46. Nestec Ltd.. Vevey/Lippincott Williams & Wilkins, Philadelphia © 2001. Human Intestinal Nutrient Transporters Ernest M. Wright Department of Physiology, UCLA School of Medicine, Los Angeles, California, USA Over the past decade, advances in molecular biology have revolutionized studies on intestinal nutrient absorption in humans. Before the advent of molecular biology, the study of nutrient absorption was largely limited to in vivo and in vitro animal model systems. This did result in the classification of the different transport systems involved, and in the development of models for nutrient transport across enterocytes (1). Nutrients are either absorbed passively or actively. Passive transport across the epithelium occurs down the nutrient's concentration gradient by simple or facilitated diffusion. The efficiency of simple diffusion depends on the lipid solubility of the nutrient in the plasma membranes—the higher the molecule's partition coefficient, the higher the rate of diffusion. Facilitated diffusion depends on the presence of simple carriers (uniporters) in the plasma membranes, and the kinetic properties of these uniporters. The rate of facilitated diffusion depends on the density, turnover number, and affinity of the uniporters in the brush border and basolateral membranes. The ' 'active'' transport of nutrients simply means that energy is provided to transport molecules across the gut against their concentration gradient. It is now well recog- nized that active nutrient transport is brought about by Na+ or H+ cotransporters (symporters) that harness the energy stored in ion gradients to drive the uphill trans- port of a solute. -

Genomic/Plasmid Dna and Rna)
NPTEL – Bio Technology – Genetic Engineering & Applications MODULE 4- LECTURE 1 ISOLATION AND PURIFICATION OF NUCLEIC ACIDS (GENOMIC/PLASMID DNA AND RNA) 4-1.1. Introduction Every gene manipulation procedure requires genetic material like DNA and RNA. Nucleic acids occur naturally in association with proteins and lipoprotein organelles. The dissociation of a nucleoprotein into nucleic acid and protein moieties and their subsequent separation, are the essential steps in the isolation of all species of nucleic acids. Isolation of nucleic acids is followed by quantitation of nucleic acids generally done by either spectrophotometric or by using fluorescent dyes to determine the average concentrations and purity of DNA or RNA present in a mixture. Isolating the genetic material (DNA) from cells (bacterial, viral, plant or animal) involves three basic steps- • Rupturing of cell membrane to release the cellular components and DNA • Separation of the nucleic acids from other cellular components • Purification of nucleic acids 4-1.2. Isolation and Purification of Genomic DNA Genomic DNA is found in the nucleus of all living cells with the structure of double- stranded DNA remaining unchanged (helical ribbon). The isolation of genomic DNA differs in animals and plant cells. DNA isolation from plant cells is difficult due to the presence of cell wall, as compared to animal cells. The amount and purity of extracted DNA depends on the nature of the cell. The method of isolation of genomic DNA from a bacterium comprises following steps (Figure 4-1.2.)- 1. Bacterial culture growth and harvest. 2. Cell wall rupture and cell extract preparation. Joint initiative of IITs and IISc – Funded by MHRD Page 1 of 57 NPTEL – Bio Technology – Genetic Engineering & Applications 3. -

Eukaryotic Expression Cloning with an Antimetastatic
[CANCER RESEARCH 59, 3812–3820, August 1, 1999] Eukaryotic Expression Cloning with an Antimetastatic Monoclonal Antibody Identifies a Tetraspanin (PETA-3/CD151) as an Effector of Human Tumor Cell Migration and Metastasis1 Jacqueline E. Testa,2,3 Peter C. Brooks,4 Jian-Min Lin,5 and James P. Quigley3 Department of Pathology, State University of New York at Stony Brook, Stony Brook, New York 11794 ABSTRACT interactions with different matrix proteins. Our efforts to identify novel metastasis-associated antigens have, therefore, focused on the tumor cell A monoclonal antibody (mAb), 50-6, generated by subtractive immuniza- surface. As our model system, we have used a highly metastatic human tion, was found to specifically inhibit in vivo metastasis of a human epider- epidermoid carcinoma cell line, HEp-3, which disseminates to host lung moid carcinoma cell line, HEp-3. The cDNA of the cognate antigen of mAb 50-6 was isolated by a modified eukaryotic expression cloning protocol from tissue. The distinct characteristics and behavior patterns of this tumor cell a HEp-3 library. Sequence analysis identified the antigen as PETA-3/CD151, line have been described by Ossowski in a series of papers published a recently described member of the tetraspanin family of proteins. The cloned between 1980 and 1998 (7–9). These cells are very aggressive and readily antigen was also recognized by a previously described antimetastatic anti- give rise to metastasizing tumors in both the chicken embryo (10–12) and body, mAb 1A5. Inhibition of HEp-3 metastasis by the mAbs could not be in the nude mouse model (13, 14). -

A Genome-Scale Protein Interaction Profile of Drosophila P53 Uncovers Additional Nodes of the Human P53 Network
A genome-scale protein interaction profile of Drosophila p53 uncovers additional nodes of the human p53 network Andrea Lunardia,b,1, Giulio Di Minina,b,1, Paolo Proveroc, Marco Dal Ferroa,b, Marcello Carottia,b, Giannino Del Sala,b, and Licio Collavina,b,2 aLaboratorio Nazionale Consorzio Interuniversitario per le Biotecnologie (LNCIB), Area Science Park, 34012 Trieste, Italy; bDipartimento di Scienze della Vita, Università degli Studi di Trieste, 34129 Trieste, Italy; and cMolecular Biotechnology Center and Dipartimento di Genetica, Biologia e Biochimica, Università degli Studi di Torino, 10126 Torino, Italy Communicated by Marc W. Kirschner, Harvard Medical School, Boston, MA, March 2, 2010 (received for review June 9, 2009) Thegenomeofthefruitfly Drosophila melanogaster contains a single to better understand the functions of all members of the p53 family p53-like protein, phylogenetically related to the ancestor of the mam- in mammals. malian p53 family of tumor suppressors. We reasoned that a compre- hensive map of the protein interaction profile of Drosophila p53 Results (Dmp53) might help identify conserved interactions of the entire Small Pool in Vitro Expression Cloning (IVEC) Screen for Drosophila p53 family in man. Using a genome-scale in vitro expression cloning p53 Interactors. The Drosophila Gene Collection (DGC) comprises approach, we identified 91 previously unreported Dmp53 interactors, full-length annotated cDNAs of the majority of known genes in considerably expanding the current Drosophila p53 interactome. D. melanogaster (16). Clones from DGC1.0 and DGC2.0 were pu- Looking for evolutionary conservation of these interactions, we rified and pooled in groups of 24. Recombinant maltose binding tested 41 mammalian orthologs and found that 37 bound to one or protein (MBP)-Dmp53 fusion protein was prepared from Bacu- more p53-family members when overexpressed in human cells. -

Expression Cloning of the Cdna for a Polypeptide Associated With
Proc. Natl. Acad. Sci. USA Vol. 92, pp. 1495-1499, February 1995 Physiology Expression cloning of the cDNA for a polypeptide associated with rat hepatic sinusoidal reduced glutathione transport: Characteristics and comparison with the canalicular transporter JLAN-Ri YI*, SHILUN LU*, JOSE FERNAINDEZ-CHECAt, AND NEIL KAPLOWITZ*1 *Division of Gastrointestinal and Liver Diseases, Department of Medicine, University of Southern California School of Medicine, Los Angeles, CA 90033; and tLiver Unit, Hospital Clinic i Provincial, Universidad de Barcelona, Barcelona 08036, Spain Communicated by Leonard A. Herzenberg, Stanford University, Stanford, CA, November 23, 1994 (received for review July 27, 1994) ABSTRACT Using the Xenopus oocyte expression system, canalicular systems are facilitative, bidirectional, and exhibit we previously identified an -4-kb fraction of mRNA from rat low affinity and high capacity (5-7). However, differences in liver that expresses sulfobromophthalein reduced glutathione certain physiological characteristics of the two low-affinity S-conjugate (BSP-GSH)-insensitive and an -2.5-kb fraction transport systems permit their functional distinction. Sinusoi- expressing BSP-GSH-sensitive reduced glutathione (GSH) dal GSH transport is cis-inhibited by sulfobromophthalein transport. From the former, a 4.05-kb cDNA was cloned and GSH S-conjugate (BSP-GSH) (7, 8), trans-inhibited by me- characterized as the putative rat canalicular GSH trans- thionine (9-11) or cystathionine (12), and unchanged after porter. Starting with a cDNA library constructed from the phenobarbital treatment (6, 13). The low-affinity canalicular -2.5-kb fraction, we have now isolated a single clone that GSH transport is not affected by these inhibitors (12, 14) but leads to expression of a BSP-GSH- and cystathionine- is induced by phenobarbital (6, 13). -
Development of a Highly Efficient Expression Cdnacloning System
Proc. Nati. Acad. Sci. USA Vol. 88, pp. 5167-5171, June 1991 Cell Biology Development of a highly efficient expression cDNA cloning system: Application to oncogene isolation (mouse hepatoma/stable expression/plasmid rescue/B-raf gene/in vivo activation) TORU MIKI*, TIMOTHY P. FLEMING*, MARCO CRESCENZI*t, CHRISTOPHER J. MOLLOY*t, SHELLEY B. BLAM*§, STEVEN H. REYNOLDS¶, AND STUART A. AARONSON*JI *Laboratory of Cellular and Molecular Biology, National Cancer Institute, Building 37, Room 1E24, Bethesda, MD 20892; and sNational Institute of Environmental Health Sciences, Biological Risk Assessment Program, Research Triangle Park, NC 27709 Communicated by G. D. Aurbach, March 19, 1991 (receivedfor review October 16, 1990) ABSTRACT We developed an expression cDNA cloning Cells were maintained in Dulbecco's modified Eagle's me- system capable of generating high-complexity libraries with dium (DMEM)/5% calf serum. unidirectionally inserted cDNA fragments and allowing effi- Plasmid Rescue. Genomic DNA (1.22g) was cleaved byXho cient plasmid rescue. As an application of this system, a cDNA I and ligated at the concentration of3 ng/,ul. Four aliquots (100 library was constructed from an NIH 3T3 transformant in- Al each) of PLK-F' competent cells (Stratagene) were trans- duced by mouse hepatocellular carcinoma DNA. Transfection formed by the ligated DNA, as directed by the manufacturer. of NIH 3T3 cells by the library DNA led to the detection of After heat shock, the cells were diluted 10-fold with S.O.C. several transformed foci from which identical plasmids with medium (BRL) containing 1 mM isopropyl /8-D-thiogalacto- transforming ability could be rescued. -
F. Katzen Et Al. the Past, Present and Future of Cell-Free Protein Synthesis
Review TRENDS in Biotechnology Vol.23 No.3 March 2005 The past, present and future of cell-free protein synthesis Federico Katzen1, Geoffrey Chang2 and Wieslaw Kudlicki1 1Invitrogen Corporation, 1600 Faraday Avenue, Carlsbad, CA 92008, USA 2Department of Molecular Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, CB-105, La Jolla, CA 92037, USA Recent technical advances have revitalized cell-free Configurations and history expression systems to meet the increasing demands In vitro translation systems are based on the early for protein synthesis. Cell-free systems offer several demonstration that cell integrity is not required for advantages over traditional cell-based expression protein synthesis to occur. In its simplest form, translation methods, including the easy modification of reaction can be accomplished using a crude lysate from any given conditions to favor protein folding, decreased sensitivity organism (that provides the translational machinery, to product toxicity and suitability for high-throughput accessory enzymes, tRNA and factors) in combination strategies because of reduced reaction volumes and with exogenously added RNA template, amino acids and process time. Moreover, improvements in translation an energy supply. This classical in vitro translation efficiency have resulted in yields that exceed a milligram scheme is called ‘uncoupled’ as opposed to the ‘coupled’ of protein per milliliter of reaction mix. We review the or ‘combined’ transcription and translation configuration advances on this expanding -

Working with Molecular Genetics Chpt. 3: Isolating and Analyzing Genes
Working with Molecular Genetics Chpt. 3: Isolating and Analyzing Genes CHAPTER 3 ISOLATING AND ANALYZING GENES Recombinant DNA, Polymerase Chain Reaction and Applications to Eukaryotic Gene Structure and Function The first two chapters covered many important aspects of genes, such as how they function in inheritance, how they code for protein (in general terms) and their chemical nature. All this was learned without having a single gene purified. A full understanding of a gene, or the entire set of genes in a genome, requires that they be isolated and then studied intensively. Once a gene is “in hand”, in principal one can determine both its biochemical structures and its function(s) in an organism. One of the goals of biochemistry and molecular genetics is to assign particular functions to individual or composite structures. This chapter covers some of the techniques commonly used to isolate genes and illustrates some of the analyses that can be done on isolated genes. Methods to purify some abundant proteins were developed early in the 20th century, and some of the experiments on the fine structure of the gene (colinearity of gene and protein for trpA and tryptophan synthase) used microbial genetics and proteins sequencing. However, methods to isolate genes were not developed until the 1960’s, and the were applicable to only a few genes. All this changed in the late 1970’s with the development of recombinant DNA technology, or molecular cloning. This technique enabled researchers to isolate any gene from any organism from which one could isolate intact DNA (or RNA). The full potential to provide access to all genes of organisms is now being realized as full genomes are sequenced. -
Basics of Molecular Biology
16 Basics of Molecular Biology Yinghui Li ([email protected]), Dingsheng Zhao State Key Laboratory of Space Medicine Fundamentals and Application, Astronaut Research and Training Center of China, Beijing 100094, China 16.1 Introduction Molecular biology is the study of biology on molecular level. The field overlaps with areas of biology and chemistry, particularly genetics and biochemistry. Molecular biology chiefly concerns itself with understanding the interactions between the various systems of a cell, including the interactions between DNA (deoxyribonucleic acid), RNA (Ribonucleic acid) and protein biosynthesis as well as learning how these interactions are regulated[1]. Researchers in molecular biology use specific techniques native to molecular biology (see the techniques section), but they combine these with techniques and ideas from genetics and biochemistry. There is not a definite line between these disciplines. Today the terms molecular biology and biochemistry are nearly interchangeable. Biochemistry is the study of chemical substances and vital processes occurring in living organisms. Biochemists focus heavily on the role, function, and structure of biomolecules. The study of chemistry behind biological processes and the synthesis of biologically active molecules are examples of biochemistry. Genetics is the study of the effects of genetic differences on organisms. Often this can be inferred by the absence of a normal component (e.g. one gene). Mutants lack one or more functional components with respect to the so-called “wild type” or normal phenotype. Genetic interactions (epistasis) can often confound simple interpretations of such “knock-out” studies. Molecular biology is the study of molecular underpinnings of the process of replication, transcription and translation of the genetic material. -

Expression Cloning of a Human Cdna Encoding Folylpoly (Gamma
Proc. Nati. Acad. Sci. USA Vol. 89, pp. 9151-9155, October 1992 Biochemistry Expression cloning of a human cDNA encoding folylpoly(y- glutamate) synthetase and determination of its primary structure TIMOTHY A. GARROW*, ARIE ADMONt, AND BARRY SHANE*t *Department of Nutritional Sciences and tHoward Hughes Medical Institute, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720 Communicated by Jesse C. Rabinowitz, June 25, 1992 ABSTRACT A human cDNA for folylpoly(y-glutamate) have been reported and pig liver FPGS has been purified to synthetase [FPGS; tetrahydrofolate:L-glutamate r-ligase (ADP homogeneity (19) but, in each case, only small amounts of forming), EC 6.3.2.17] has been cloned by functional comple- protein have been obtained, which has limited characteriza- mentation of an Escherichia coli foiC mutant. The cDNA tion to kinetic analyses (15-22). The low abundance and encodes a 545-residue protein of Mr 60,128. The deduced instability of mammalian FPGS have complicated its purifi- sequence has regions that are highly homologous to peptide cation in sufficient quantity to carry out mechanistic studies. sequences obtained from purified pig liver FPGS and shows CHO-human (23) and CHO-mouse (24) hybrids have been limited homology to the E. coli and Lactobacilus casei FPGSs. used to localize the human and murine FPGS genes to Expression ofthe cDNA in E. coli results in elevated expression chromosomes 9 and 2, respectively. To aid in the further of an enzyme with characteristics of mammalian FPGS. Ex- characterization of the mammalian protein and to study its pression of the cDNA in AUXB1, a mammalian cell lacking regulation, we initially attempted to clone the human FPGS FPGS activity, overcomes the cell's requirement for thymidine gene by purifying human sequences capable of complement- and purines but does not overcome the cell's glycine auxotro- ing CHO AUXB1 cells to the wild-type phenotype using phy, consistent with expression of the protein in the cytosol but multiple rounds of DNA transfection (25).