Molecular Phylogeny, Evolution Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

San Gabriel Chestnut ESA Petition
BEFORE THE SECRETARY OF THE INTERIOR PETITION TO THE U.S. FISH AND WILDLIFE SERVICE TO PROTECT THE SAN GABRIEL CHESTNUT SNAIL UNDER THE ENDANGERED SPECIES ACT © James Bailey CENTER FOR BIOLOGICAL DIVERSITY Notice of Petition Ryan Zinke, Secretary U.S. Department of the Interior 1849 C Street NW Washington, D.C. 20240 [email protected] Greg Sheehan, Acting Director U.S. Fish and Wildlife Service 1849 C Street NW Washington, D.C. 20240 [email protected] Paul Souza, Director Region 8 U.S. Fish and Wildlife Service Pacific Southwest Region 2800 Cottage Way Sacramento, CA 95825 [email protected] Petitioner The Center for Biological Diversity is a national, nonprofit conservation organization with more than 1.3 million members and supporters dedicated to the protection of endangered species and wild places. http://www.biologicaldiversity.org Failure to grant the requested petition will adversely affect the aesthetic, recreational, commercial, research, and scientific interests of the petitioning organization’s members and the people of the United States. Morally, aesthetically, recreationally, and commercially, the public shows increasing concern for wild ecosystems and for biodiversity in general. 1 November 13, 2017 Dear Mr. Zinke: Pursuant to Section 4(b) of the Endangered Species Act (“ESA”), 16 U.S.C. §1533(b), Section 553(3) of the Administrative Procedures Act, 5 U.S.C. § 553(e), and 50 C.F.R. §424.14(a), the Center for Biological Diversity and Tierra Curry hereby formally petition the Secretary of the Interior, through the United States Fish and Wildlife Service (“FWS”, “the Service”) to list the San Gabriel chestnut snail (Glyptostoma gabrielense) as a threatened or endangered species under the Endangered Species Act and to designate critical habitat concurrently with listing. -

New Design Water Treatment Plant /'7I.E.C
Final Report Potomac River Source Water ~s.ejgitllm~tsjjK.,..~IW1I~ for Marylan . Prepared by: Becker and O'Melia, LLC in association with Straughan Environmental Services, Inc. Becker and O'Melia, LLC WATER PROCESS RESEARCHERS AND CONSULTANTS POTOMAC SOURCE WATER ASSESSMENTS Introduction The 1996 Safe Drinking Water Act Amendments required states to develop and implement source water assessment prograIlli to evaluate the safety of all public drinking water systems. A Source Water Assessment (SWA) is a process for evaluating the vulnerability to contamination ofthe source of a public drinking water supply. The assessment does not address the treatment processes, or the storage and distribution aspects of the water system, which are covered under separate provisions of the Safe Drinking Water Act. The Maryland Department of the Environment (MDE) is the lead state agency in this source water assessment effort. There are three main steps in the assessment process: (1) delineating the watershed drainage area that is likely to contribute to the drinking water supply, (2) identijYing potential contaminants within that area and (3) assessing the vulnerability, or susceptibility, of the system to those contaminants. Public notification is the final component of the source water assessment report process. The goal of the source water assessment program is to provide a framework for local stakeholders and governments in developing a Source Water Protection Plan. The source water assessments for Maryland water systems utilizing the Potomac River was undertaken as a joint effort by MDE, the Washington Suburban Sanitary Conunission (WSSC), and several consultants, including The Center for Watershep Ptotection, and Becker & O'Melia, the lead consultant. -

NON-TIDAL BENTHIC MONITORING DATABASE: Version 3.5
NON-TIDAL BENTHIC MONITORING DATABASE: Version 3.5 DATABASE DESIGN DOCUMENTATION AND DATA DICTIONARY 1 June 2013 Prepared for: United States Environmental Protection Agency Chesapeake Bay Program 410 Severn Avenue Annapolis, Maryland 21403 Prepared By: Interstate Commission on the Potomac River Basin 51 Monroe Street, PE-08 Rockville, Maryland 20850 Prepared for United States Environmental Protection Agency Chesapeake Bay Program 410 Severn Avenue Annapolis, MD 21403 By Jacqueline Johnson Interstate Commission on the Potomac River Basin To receive additional copies of the report please call or write: The Interstate Commission on the Potomac River Basin 51 Monroe Street, PE-08 Rockville, Maryland 20850 301-984-1908 Funds to support the document The Non-Tidal Benthic Monitoring Database: Version 3.0; Database Design Documentation And Data Dictionary was supported by the US Environmental Protection Agency Grant CB- CBxxxxxxxxxx-x Disclaimer The opinion expressed are those of the authors and should not be construed as representing the U.S. Government, the US Environmental Protection Agency, the several states or the signatories or Commissioners to the Interstate Commission on the Potomac River Basin: Maryland, Pennsylvania, Virginia, West Virginia or the District of Columbia. ii The Non-Tidal Benthic Monitoring Database: Version 3.5 TABLE OF CONTENTS BACKGROUND ................................................................................................................................................. 3 INTRODUCTION .............................................................................................................................................. -

Program Overview
WWeett WWaaddeerrss aanndd BBeeyyoonndd TThhee CCoonnddiittiioonn ooff OOuurr SSttaattee’’ss WWaatteerrss AA CCiittiizzeenn’’ss PPeerrssppeeccttiivvee 1 WV Department of Environmental Protection Division of Water and Waste Management, Nonpoint Section 601 57th Street, SE Charleston, WV 25304 The document was prepared by Tim Craddock, WV DEP’s Citizens’ Monitoring Coordinator and is available electronically in Portable Document Format (PDF). To request your copy send e-mail to Tim Craddock at: [email protected]. ACKNOWLEDGEMENTS Color photographs provided by: Alana Hartman, DEP’s Potomac Basin Coordinator; Abby Chappel, WV River Network; Sherry Evasic, Blue Heron Environmental Network; Neil Gillies, Cacapon Institute; Suzanne Hubbard, The Mountain Institute; Renee Cain, Lower West Fork Watershed Association; Martin Christ, Friends of Deckers Creek; Bobby Bonnett, Heizer-Manila Watershed Organization; Diana Green, Davis Creek Watershed Association; James Grey, Morris Creek Watershed Association; Larry Orr, Kanawha Valley Chapter of Trout Unlimited; Valerie Wilson, Science Teacher, Oak Hill Catholic Center; Brad Durst, WV Conservation Agency and Curtis Canada, Upper Guyandotte Watershed Association. WV Save Our Streams would like to recognize all the volunteer monitors, not only those directly associated with the program, but any others who have given their time and energy in an effort to protect our state’s streams and rivers. WV Save Our Streams would also like to recognize all of the agency and other partners who have provided assistance of any kind, to help guide volunteers through the myriad of processes involved with water quality issues. “Perception is not acquired by formal education, nor is it reserved for persons learned in the arts or sciences. -

Diseases of Trees in the Great Plains
United States Department of Agriculture Diseases of Trees in the Great Plains Forest Rocky Mountain General Technical Service Research Station Report RMRS-GTR-335 November 2016 Bergdahl, Aaron D.; Hill, Alison, tech. coords. 2016. Diseases of trees in the Great Plains. Gen. Tech. Rep. RMRS-GTR-335. Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station. 229 p. Abstract Hosts, distribution, symptoms and signs, disease cycle, and management strategies are described for 84 hardwood and 32 conifer diseases in 56 chapters. Color illustrations are provided to aid in accurate diagnosis. A glossary of technical terms and indexes to hosts and pathogens also are included. Keywords: Tree diseases, forest pathology, Great Plains, forest and tree health, windbreaks. Cover photos by: James A. Walla (top left), Laurie J. Stepanek (top right), David Leatherman (middle left), Aaron D. Bergdahl (middle right), James T. Blodgett (bottom left) and Laurie J. Stepanek (bottom right). To learn more about RMRS publications or search our online titles: www.fs.fed.us/rm/publications www.treesearch.fs.fed.us/ Background This technical report provides a guide to assist arborists, landowners, woody plant pest management specialists, foresters, and plant pathologists in the diagnosis and control of tree diseases encountered in the Great Plains. It contains 56 chapters on tree diseases prepared by 27 authors, and emphasizes disease situations as observed in the 10 states of the Great Plains: Colorado, Kansas, Montana, Nebraska, New Mexico, North Dakota, Oklahoma, South Dakota, Texas, and Wyoming. The need for an updated tree disease guide for the Great Plains has been recog- nized for some time and an account of the history of this publication is provided here. -

Annual Report 2010–2011
Australian Museum 2010–2011 Report Annual Australian Museum Annual Report 2010–2011 Australian Museum Annual Report 2010 – 2011 ii Australian Museum Annual Report 2010 –11 The Australian Museum Annual Report 2010 –11 Availability is published by the Australian Museum Trust, This annual report has been designed for accessible 6 College Street Sydney NSW 2010. online use and distribution. A limited number of copies have been printed for statutory purposes. © Australian Museum Trust 2011 This report is available at: ISSN 1039-4141 www.australianmuseum.net.au/Annual-Reports. Editorial Further information on the research and education Project management: Wendy Rapee programs and services of the Australian Museum Editing and typesetting: Brendan Atkins can be found at www.australianmuseum.net.au. Proofreading: Lindsay Taaffe Design and production: Australian Museum Environmental responsibility Design Studio Printed on Sovereign Offset, an FSC- certified paper from responsibly grown fibres, made under an All photographs © Australian Museum ISO 14001– accredited environmental management 2011, unless otherwise indicated. system and without the use of elemental chlorine. Contact Australian Museum 6 College Street Sydney NSW 2010 Open daily 9.30 am – 5.00 pm t 02 9320 6000 f 02 9320 6050 e [email protected] w www.australianmuseum.net.au www.facebook.com/australianmuseum www.twitter.com/austmus www.youtube.com/austmus front cover: The Museum's after-hours program, Jurassic Lounge, attracted a young adult audience to enjoy art, music and new ideas. Photo Stuart Humphreys. iii Minister The Hon. George Souris, MP and Minister for the Arts Governance The Museum is governed by a Trust established under the Australian Museum Trust Act 1975. -

Malacologica
FOLIA Folia Malacol. 24(3): 111–177 MALACOLOGICA ISSN 1506-7629 The Association of Polish Malacologists Faculty of Biology, Adam Mickiewicz University Bogucki Wydawnictwo Naukowe Poznań, September 2016 http://dx.doi.org/10.12657/folmal.024.008 PATTERNS OF SPATIO-TEMPORAL VARIATION IN LAND SNAILS: A MULTI-SCALE APPROACH SERGEY S. KRAMARENKO Mykolaiv National Agrarian University, Paryzka Komuna St. 9, Mykolaiv, 54020, Ukraine (e-mail: [email protected]) ABSTRACT: Mechanisms which govern patterns of intra-specific vatiation in land snails were traced within areas of different size, using Brephulopsis cylindrica (Menke), Chondrula tridens (O. F. Müller), Xeropicta derbentina (Krynicki), X. krynickii (Krynicki), Cepaea vindobonensis (Férussac) and Helix albescens Rossmässler as examples. Morphometric shell variation, colour and banding pattern polymorphism as well as genetic polymorphism (allozymes and RAPD markers) were studied. The results and literature data were analysed in an attempt to link patterns to processes, with the following conclusions. Formation of patterns of intra- specific variation (initial processes of microevolution) takes different course at three different spatial scales. At micro-geographical scale the dominant role is played by eco-demographic characteristics of the species in the context of fluctuating environmental factors. At meso-geographical scale a special part is played by stochastic population-genetic processes. At macro-geographical scale more or less distinct clinal patterns are associated with basic macroclimatic -
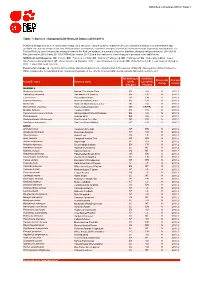
Table 7: Species Changing IUCN Red List Status (2010-2011)
IUCN Red List version 2011.2: Table 7 Table 7: Species changing IUCN Red List Status (2010-2011) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2010 (IUCN Red List version 2010.4) and 2011 (IUCN Red List version 2011.2) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered, EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.) IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2010) List (2011) change version Category Category MAMMALS Bradypus torquatus Maned Three-toed Sloth EN VU N 2011.1 Callicebus oenanthe San Martin Titi Monkey EN CR N 2011.1 Equus ferus Przewalski's Horse CR EN G 2011.2 -

An Inventory of the Land Snails and Slugs (Gastropoda: Caenogastropoda and Pulmonata) of Knox County, Tennessee Author(S): Barbara J
An Inventory of the Land Snails and Slugs (Gastropoda: Caenogastropoda and Pulmonata) of Knox County, Tennessee Author(s): Barbara J. Dinkins and Gerald R. Dinkins Source: American Malacological Bulletin, 36(1):1-22. Published By: American Malacological Society https://doi.org/10.4003/006.036.0101 URL: http://www.bioone.org/doi/full/10.4003/006.036.0101 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Amer. Malac. Bull. 36(1): 1–22 (2018) An Inventory of the Land Snails and Slugs (Gastropoda: Caenogastropoda and Pulmonata) of Knox County, Tennessee Barbara J. Dinkins1 and Gerald R. Dinkins2 1Dinkins Biological Consulting, LLC, P O Box 1851, Powell, Tennessee 37849, U.S.A [email protected] 2McClung Museum of Natural History and Culture, 1327 Circle Park Drive, Knoxville, Tennessee 37916, U.S.A. Abstract: Terrestrial mollusks (land snails and slugs) are an important component of the terrestrial ecosystem, yet for most species their distribution is not well known. -

Entre Los Stylommatophora (Mollusca: Gastropoda)
Rev. peru. biol. 16(1): 051- 056 (Agosto 2009) © Facultad de Ciencias Biológicas UNMSM Posición evolutiva de BOSTRYX y SCUTALUS dentroVersión de Online los Stylommatophora ISSN 1727-9933 Posición evolutiva de caracoles terrestres peruanos (Orthalicidae) entre los Stylommatophora (Mollusca: Gastropoda) Evolutionary position of Peruvian land snails (Orthalicidae) among Stylommatophora (Mollusca: Gastropoda) Jorge Ramirez1,2, Rina Ramírez1,2, Pedro Romero1,2, Ana Chumbe1,2, Pablo Ramírez3 1Laboratorio de Sistemática Mole- cular y Filogeografía, Facultad de Resumen Ciencias Biológicas, Universidad Nacional Mayor de San Marcos. Los géneros Bostryx y Scutalus (Orthalicidae: Bulimulinae) son endémicos de América del Sur y están principal- Email Jorge Ramirez: jolobio@ mente distribuidos en la vertiente occidental de los Andes del Perú. El objetivo del presente trabajo fue evaluar hotmail.com su posición evolutiva dentro de los gastrópodos Stylommatophora basada en el marcador mitocondrial 16S 2Departamento de Malacología y Carcinología, Museo de Historia rRNA. Fueron obtenidas cuatro secuencias las que, junto con 28 de otros Stylommatophora disponibles en el Natural, Universidad Nacional GenBank, fueron alineadas con ClustalX. La reconstrucción filogenética se realizó mediante los métodos de Mayor de San Marcos. Neighbor-Joining, Máxima Parsimonia, Máxima Verosimilitud e Inferencia Bayesiana. El alineamiento resultó en Av. Arenales 1256, Apartado 14- 371 sitios, con presencia de indels. Los dos géneros de la Familia Orthalicidae por primera vez incluidos en una 0434, Lima-14, Perú. Email Rina filogenia molecular (Bostryx y Scutalus), formaron un grupo monofilético con otro miembro de la superfamilia Ramírez: [email protected] Orthalicoidea (Placostylus), tal como lo obtenido con marcadores nucleares. Se discute también su relación 3Laboratorio de Microbiología Molecular, Facultad de Ciencias evolutiva con otros caracoles terrestres. -

Section IV – Guideline for the Texas Priority Species List
Section IV – Guideline for the Texas Priority Species List Associated Tables The Texas Priority Species List……………..733 Introduction For many years the management and conservation of wildlife species has focused on the individual animal or population of interest. Many times, directing research and conservation plans toward individual species also benefits incidental species; sometimes entire ecosystems. Unfortunately, there are times when highly focused research and conservation of particular species can also harm peripheral species and their habitats. Management that is focused on entire habitats or communities would decrease the possibility of harming those incidental species or their habitats. A holistic management approach would potentially allow species within a community to take care of themselves (Savory 1988); however, the study of particular species of concern is still necessary due to the smaller scale at which individuals are studied. Until we understand all of the parts that make up the whole can we then focus more on the habitat management approach to conservation. Species Conservation In terms of species diversity, Texas is considered the second most diverse state in the Union. Texas has the highest number of bird and reptile taxon and is second in number of plants and mammals in the United States (NatureServe 2002). There have been over 600 species of bird that have been identified within the borders of Texas and 184 known species of mammal, including marine species that inhabit Texas’ coastal waters (Schmidly 2004). It is estimated that approximately 29,000 species of insect in Texas take up residence in every conceivable habitat, including rocky outcroppings, pitcher plant bogs, and on individual species of plants (Riley in publication). -

A Phylogenetic Analysis of Polygyridae (Gastropoda: Pulmonata) Based on Mitochondrial DNA Sequence Data Nicholas A
Southern Illinois University Carbondale OpenSIUC Honors Theses University Honors Program 2012 A Phylogenetic Analysis of Polygyridae (Gastropoda: Pulmonata) Based on Mitochondrial DNA Sequence Data Nicholas A. Defreitas Southern Illinois University Carbondale, [email protected] Follow this and additional works at: http://opensiuc.lib.siu.edu/uhp_theses Recommended Citation Defreitas, Nicholas A., "A Phylogenetic Analysis of Polygyridae (Gastropoda: Pulmonata) Based on Mitochondrial DNA Sequence Data" (2012). Honors Theses. Paper 348. This Dissertation/Thesis is brought to you for free and open access by the University Honors Program at OpenSIUC. It has been accepted for inclusion in Honors Theses by an authorized administrator of OpenSIUC. For more information, please contact [email protected]. A Phylogenetic Analysis of Polygyridae (Gastropoda: Pulmonata) Based on Mitochondrial DNA Sequence Data Nicholas Defreitas University Honors Program Senior Thesis Introduction Despite the increasing use of molecular methods to determine evolutionary relationships among taxa, molecular sequence data have never been used to assess the relationships among the polygyrid snails (Gastropoda:Pulmonata:Polygyridae). This is surprising, considering how large, charismatic and common they are. Polygyrids range across North America, going as far north as parts of Canada and south as Mexico and even deeper into Central America (Pilsbry 1940). There is a particular concentration of these snails in the Appalachian Mountains, where they primarily serve as detritivores and prey for various woodland vertebrates in forest habitats. Yet despite the broad geographic distribution and high abundance of polygyrids in many forest habitats, there is still little known about their phylogeny (evolutionary relationships). Polygyrids are broadly distributed across North America. Mesodontini and Triodopsini are both found in eastern North America (Hubricht 1985).