Lassa Virus in the Host Rodent Mastomys Natalensis Within Urban Areas of N’Zerekore, Guinea
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
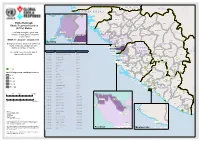
Etc Status with 21Confcase 1.Pdf
SE N E G A L M A L I GU IN EA -B IS SA U Koundara Mali ETC"-GIN-003 Koubia Gaoual Lelouma Dinguiraye Siguiri Ebola Outbreak: Labe Tougue Ebola Treatment Centres Telimele (ETCs) Status Dalaba Kouroussa Boke This Map shows the status and ETC"-GIN-012 Pita Mandiana Boffa Dabola location of each Ebola Treatment Mamou Fria Center (ETC). ETC"-GIN-001 ETC"-GIN-018 Dubreka Kindia Faranah ETC-GIN-015 WEEK 17: 20 April - 26 April 2015 " Kankan Conakry Copyright:© 2014 Esri Background colour show new confirmed Koinadugu ETC-GIN-00C3 oyah G U I N E A " Bombali cases for the last 21 days for each ConakryETC-GIN-012 district, prefecture or county. ETC"-EGT"ICN"-G00IN1-018 S I E R R A Kissidougou CÔ TE ETC-GIN-017 ETC CODE Site Name Country Forecariah " L E O N E Kerouane D' IV OI R E The number over the ETC sign is ETC-GIN-001 Conakry Region Guinea Beyla Freetownreferenced in the table. ETC-GIN-003 Kindia Region, Coyah Prefecture Guinea Kambia ETC"-SLE-034 ETC-SLE-008 ETC-GIN-007 Nzérékoré Region Guinea " Kono Gueckedou ETC-GIN-009 Nzérékoré Region Guinea Port Loko EETTCC--SSLLEE--00002571 ETC-SLE-022 "" " ETC-GIN-009 ETC-GIN-010 Nzérékoré Region Guinea ETC-SLE-031 " " ETC-GIN-010 Western " ETC-GIN-011 ETC-GIN-011 Nzérékoré Region Guinea Area Urban ETECT-SCL-SEL-0E2-4028 Tonkolili " ETC-LBR-011 ETC-GIN-012 Conakry Region Guinea "E"ETTC"C--SSLLEE--00121375 ETECT"-CS-LSEL-E0-2303226 " ETC-GIN-015 Kindia Guinea ET"C"-SLE-0116 Lofa Macenta Western " " ETC-SLE-009 ETC-GIN-017 Forecariah Guinea Area Rural " ETC-GIN-018 Conakry Guinea ETC-SLE-004 Nzerekore -

PRADD II Guinea Impact Evaluation Design Report
EVALUATION, RESEARCH AND COMMUNICATION (ERC) Property Rights and Artisanal Diamond Development Project II (PRADD II) Impact Evaluation Design Report AUGUST 2014 This document was produced for review by the United States Agency for International Development. It was prepared by Cloudburst Consulting Group, Inc. for the Evaluation, Research, and Communication (ERC) Task Order under the Strengthening Tenure and Resource Rights (STARR) IQC. Written and prepared by Heather Huntington, Michael McGovern, and Darrin Christensen. Prepared for the United States Agency for International Development, USAID Contract Number AID- OAA-TO-13-00019, Evaluation, Research and Communication (ERC) Task Order under Strengthening Tenure and Resource Rights (STARR) IQC No. AID-OAA-I-12-00030. Implemented by: Cloudburst Consulting Group, Inc. 8400 Corporate Drive, Suite 550 Landover, MD 20785-2238 EVALUATION, RESEARCH AND COMMUNICATION (ERC) Property Rights and Artisanal Diamond Development Project II (PRADD II) Impact Evaluation Design Report AUGUST 2014 DISCLAIMER The authors' views expressed in this publication do not necessarily reflect the views of the United States Agency for International Development or the United States Government. CONTENTS 36T36TCONTENTS36T36T ............................................................................................................................ 4 36T36TACRONYMS AND ABBREVIATIONS36T36T ..................................................................................... 5 36T36T1.0 INTRODUCTION36T36T .............................................................................................................. -

GSV2.1 LSC Report BISS V2 August 2011
GOLD STANDARD LOCAL STAKEHOLDER CONSULTATION REPORT CONTENTS A. Project Description 1. Project eligibility under Gold Standard 2. Current project status B. Design of Stakeholder Consultation Process 1. Description of physical meeting(s) i. Agenda ii. Non-technical summary iii. Invitation tracking table iv. Text of individual invitations v. Text of public invitations 2. Description of other consultation methods used C. Consultation Process 1. Participants’ in physical meeting(s) i. List ii. Evaluation forms 2. Pictures from physical meeting(s) 3. Outcome of consultation process i. Minutes of physical meeting(s) ii. Minutes of other consultations iii. Assessment of all comments iv. Revisit sustainable development assessment v. Summary of changes to project design based on comments D. Sustainable Development Assessment 1. Own sustainable development assessment i. ‘Do no harm’ assessment ii. Sustainable development matrix 2. Stakeholders blind sustainable development matrix 3. Consolidated sustainable development matrix E. Discussion on Sustainability Monitoring Plan F. Description of Stakeholder Feedback Round Annex 1. Original participants list Annex 2. Original feedback forms Annex 3. Original non-technical summary SECTION A. PROJECT DESCRIPTION A. 1. Project eligibility under the Gold Standard The efficient cook stove project in Guinea falls under the “End-use Energy Efficiency Improvement” category as mentioned in the GS Toolkit Annexes. The project will generate an annual average GHG emissions reduction volume around 8000 teqCO2. According to the Gold Standard classification, the Project is qualified as a “small scale project”. A. 2. Current project status General description of the project: The purpose of the project is to improve conditions of Guinean households in Kindia area (Republic of Guinea) and fight against global warming and deforestation by promoting the use of an efficient cook stove (vernacular name: « kolpot fötönkanté »). -

Republic of Guinea: Overcoming Growth Stagnation to Reduce Poverty
Report No. 123649-GN Public Disclosure Authorized REPUBLIC OF GUINEA OVERCOMING GROWTH STAGNATION TO REDUCE POVERTY Public Disclosure Authorized SYSTEMATIC COUNTRY DIAGNOSTIC March 16, 2018 International Development Association Country Department AFCF2 Public Disclosure Authorized Africa Region International Finance Corporation Sub-Saharan Africa Department Multilateral Investment Guarantee Agency Sub-Saharan Africa Department Public Disclosure Authorized WORLD BANK GROUP IBRD IFC Regional Vice President: Makhtar Diop : Vice President: Dimitris Tsitsiragos Country Director: Soukeyna Kane Director: Vera Songwe : Country Manager: Rachidi Radji Country Manager: Cassandra Colbert Task Manager: Ali Zafar : Resident Representative: Olivier Buyoya Co-Task Manager: Yele Batana ii LIST OF ACRONYMS AGCP Guinean Central Procurement Agency ANASA Agence Nationale des Statistiques Agricoles (National Agricultural Statistics Agency) Agence de Promotion des Investissements et des Grands Travaux (National Agency for APIX Promotion of Investment and Major Works) BCRG Banque Centrale de la République de Guinée (Central Bank of Guinea) CEQ Commitment to Equity CGE Computable General Equilibrium Conseil National pour la Démocratie et le Développement (National Council for CNDD Democracy and Development) Confédération Nationale des Travailleurs de Guinée (National Confederation of CNTG Workers of Guinea) CPF Country Partnership Framework CPIA Country Policy and Institutional Assessment CRG Crédit Rural de Guinée (Rural Credit of Guinea) CWE China Water and -

2.3.7 Guinea Border Crossing of Madina Oula
2.3.7 Guinea Border Crossing of Madina Oula Overview Daily Capacity Customs Clearance Other Relevant Information Overview Madina-Oula is a town and sub-prefecture in the Kindia Prefecture in the Kindia Region of western Guinea. The road from Madina Oula to Kindia is not paved and can deteriorate during the rainy season. BORDER CROSSING LOCATION & CONTACT Name of Border Crossing Madina Oula [Guinea] Saina [Sierra Leone] Province or District Kindia Nearest Town or City Kindia 65 km Latitude 9.880611 Longitude -12.4475 Managing Authority/Agency Customs Authority Contact Person N/A Travel Times Nearest International Airport Conakry International Airport 186 km Truck: 5 hours Car: 3 hours Nearest Port Port Autonome de Conakry 196 km Truck: 5 hours Car: 3 hours Nearest Major Market Kindia 65 km Truck: 2 hours Car: 1 hour Other Information There are no weighing bridges en-route. Fueling stations are available in nearest towns. Hours of Operation MONDAYS 0800 - 1830 TUESDAYS 0800 - 1830 WEDNESDAYS 0800 - 1830 Page 1 THURSDAYS 0800 - 1830 FRIDAYS 0800 - 1830 SATURDAYS 0800 - 1830 SUNDAYS 0800 - 1830 NATIONAL HOLIDAYS No closing days. SEASONAL CONSTRAINTS Rainy season might make the access to the border difficult and delays might occur. Daily Capacity The borders were closed during the Ebola outbreak in 2014/2015. Private cars are not provided a separate lane. Customs Clearance In order to obtain a customs clearance, all the documents should be prepared and approved by the Customs authority in Conakry, then transmitted to the regional customs office respectively. A copy should be made available at the border post by the requester/transporter. -

GUINEA Ebola Situation Report
GUINEA Ebola Situation Report 29 April 2015 HIGHLIGHTS SITUATION IN NUMBERS As of 29 APRIL 2015 The total number of confirmed cases of Ebola rose to 3,158 this week, according to WHO’s Epidemiological Situation Report. The total number of confirmed, suspected and probable cases 3,584 climbed to 3,584. Cases of Ebola (3,158 confirmed) The number of confirmed deaths from Ebola rose to 1,964, with a total of 2,379 confirmed, suspected and probable deaths. 2,379 Unfortunately, some cases continue to be identified after post Deaths (1,964 confirmed) mortem testing or arisen from non-contacts, suggesting the need to reinforce surveillance and community engagement. 596 The Government-led four-day emergency health campaign with Cases among children 0-17 partners continued this week from 24 to 27 April 2015 in the (confirmed) prefecture of Coyah to end the Ebola outbreak. UNICEF and partners organized community and health workers into 610 teams reaching over 286,314 people (57,267 households) with 349 information on how to protect themselves against the disease Deaths of children and youth aged and provided them with soap. 0-17 (confirmed) Since the beginning of the Ebola outbreak, UNICEF constructed and rehabilitated more than 209 water points in the most affected regions of Guinea. This brings the total number of people 4,350,633 with improved access to water to more than 62,700. In 2015, Children in affected areas more than 250 water points are planned to be constructed or rehabilitated. This will be the highest result achieved under the UNICEF Guinea Country Programme 2013-2017 aiming at drilling 187 or rehabilitating 400 boreholes. -

Mapping Maternal and Newborn Healthcare Access in West African Countries
Mapping maternal and newborn healthcare access in West African Countries Dorothy Ononokpono University of Uyo Bernard Baffour ( [email protected] ) Australian National University https://orcid.org/0000-0002-9820-2617 Alice Richardson Australian National University Research article Keywords: Maternal and newborn health, districts, West Africa, mapping, geospatial analysis, buffer analysis. Posted Date: August 13th, 2019 DOI: https://doi.org/10.21203/rs.2.11583/v2 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/28 Abstract Background: The Sustainable Development Goal (SDG) three emphasizes the need to improve maternal and newborn health, and reduce global maternal mortality ratio to less than 70 per 100 000 live births by 2030. Achieving the SDG goal 3.1 target will require evidence based data on concealed inequities in the distribution of maternal and child health outcomes and their linkage to healthcare access. The objectives of this study were to estimate the number of women of reproductive age, pregnancies and live births at subnational level using high resolution maps and to quantify the number of pregnancies within user- dened distances or travel times of a health facility in three poor resource West African countries: Mali, Guinea and Liberia. Methods: The maternal and newborn health outcomes were estimated and mapped for the purpose of visualization using geospatial analytic tools. Buffer analysis was then performed to assess the proximity of pregnancies to health facilities with the aim of identifying pregnancies with inadequate access (beyond 50km) to a health facility. Results: Results showed wide variations in the distribution of maternal and newborn health outcomes across the countries of interest and districts of each of the countries. -

Guinea Staple Food Market Fundamentals March 2017
GUINEA STAPLE FOOD MARKET FUNDAMENTALS MARCH 2017 This publication was produced for review by the United States Agency for International Development. It was prepared by Chemonics International Inc. for the Famine Early Warning Systems Network (FEWS NET), contract number AID-OAA-I-12-00006. The authors’ views expressed in this publication do not necessarily reflect the views of the United States Agency for International Development or the United States government. FEWS NET Guinea Staple Food Market Fundamentals 2017 About FEWS NET Created in response to the 1984 famines in East and West Africa, the Famine Early Warning Systems Network (FEWS NET) provides early warning and integrated, forward-looking analysis of the many factors that contribute to food insecurity. FEWS NET aims to inform decision makers and contribute to their emergency response planning; support partners in conducting early warning analysis and forecasting; and provide technical assistance to partner-led initiatives. To learn more about the FEWS NET project, please visit http://www.fews.net Disclaimer This publication was prepared under the United States Agency for International Development Famine Early Warning Systems Network (FEWS NET) Indefinite Quantity Contract, AID-OAA-I-12-00006. The authors’ views expressed in this publication do not necessarily reflect the views of the United States Agency for International Development or the United States government. Acknowledgements FEWS NET gratefully acknowledges the network of partners in Guinea who contributed their time, analysis, and data to make this report possible. See the list of participating organizations in Annex 2. Market Fundamentals Workshop Participants. Famine Early Warning Systems Network i FEWS NET Guinea Staple Food Market Fundamentals 2017 Table of Contents Executive Summary .................................................................................................................................................................... -

Investment Opportunities in Agriculture Guinea
SECTION: INVESTMENT OPPORTUNITIES IN AGRICULTURE GUINEA Project title: Constructing an agro-industrial starch processing complex Name of organization: Ministère de l'Industrie (Ministry of Industry)/APIP (Agence de promotion des investissements privés, agency for the promotion of private investments) Year of establishment: 2015 Project location: Timbi Madina, Pita Prefecture, Mamou Region, Guinea Project type: Greenfield Investment type: PPP Project status: Feasibility completed Investment amount required (EUR €): 13,000,000.00 This project has been developed by the Ministry of Industry and the Agency for the Promotion of Private Investments of Guinea to construct an agro-industrial complex for processing potatoes and manioc. The complex will have three components: a bio-reactor, to generate electricity, high-quality fertilizer and water for irrigation in the off-season; a plant for the extraction of starch and glucose; and a facility for the manufacture of monosodium glutamate. There is also potential for the manufacture of additional products, including biogas, bio-degradable plastics, and bouillon cubes. Guinean farmers have had great success producing potatoes and manioc using traditional, renewable agricultural methods, but have struggled to find a market for their produce. As such, there is an opportunity to turn the excess potato and manioc crops into products for which there is a higher local demand. The developer seeks a partner with technical expertise in managing starch processing units, specifically a manufacturer of potato-processing factory equipment. The developer likewise seeks investors interested in Guinea’s agricultural sector and supporting the establishment of an agro- industrial plant with far-reaching positive impacts on small farmers in rural areas of the country by providing access to new and expanded markets. -

Citizens' Involvement in Health Governance
CITIZENS’ INVOLVEMENT IN HEALTH GOVERNANCE (CIHG) Endline Data Collection Final Report September 2020 This report was prepared with funds provided by the U.S. Agency for International Development under Cooperative Agreement AID-675-LA-17-00001. The opinions expressed herein are those of the author(s) and do not necessarily reflect the views of the U.S. Agency for International Development. Contents Executive Summary ...................................................................................... 1 I. Introduction ............................................................................................... 5 Overview ...................................................................................................... 5 Background................................................................................................... 5 II. Methodology ............................................................................................ 6 Approach ...................................................................................................... 6 Data Collection ............................................................................................. 7 Analysis ....................................................................................................... 10 Limitations .................................................................................................. 10 Safety and Security ..................................................................................... 11 III. Findings ................................................................................................ -
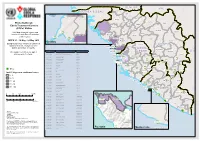
Etc Status with 21Days Confirm
SE NE GA L MA LI GU IN EA- BIS SAU Koundara Mali ETC"-GIN-003 Koubia Gaoual Lelouma Dinguiraye Siguiri Ebola Outbreak: Labe Tougue Ebola Treatment Centres Telimele (ETCs) Status Dalaba Kouroussa Boke This Map shows the status and ETC"-GIN-012 Pita Mandiana Boffa Dabola location of each Ebola Treatment Mamou Fria Center (ETC). ETC"-GIN-001 ETC"-GIN-018 Dubreka Kindia Faranah ETC-GIN-015 WEEK 21 : 18 May - 24 May 2015 " Kankan Conakry Copyright:© 2014 Esri Background colour show new confirmed Koinadugu ETC-GIN-00C3 oyah GU I N E A cases for the last 21 days for each " Bombali ConakryETC-GIN-012 district, prefecture or county. ETC"-EGT"ICN"--G00IN1 -018 SI E R R A Kissidougou CÔTE ETC-GIN-017 ETC CODE Site Name Country Forecariah " Kerouane D'IV OIR E The number over the ETC sign is LE ON E ETC-GIN-001 Conakry Region Guinea Beyla referenced in the table. ETC-SLE-034 Freetown ETC-GIN-003 Kindia Region, Coyah Prefecture Guinea Kambia " ETC-SLE-008 ETC-GIN-007 Nzérékoré Region Guinea " Kono Gueckedou ETC-GIN-009 Nzérékoré Region Guinea Port Loko EETTCC--SSLLEE--00020517 ETC-SLE-022 " " ETC-GIN-009 ETC-GIN-010 Nzérékoré Region Guinea ETC-SLE-031 " " ETC-GIN-010 Western " ETC-GIN-011 ETC-GIN-011 Nzérékoré Region Guinea Area Urban ETCET-SCL-ES-L0E2-4028 Tonkolili " ETC-LBR-011 ETC-GIN-012 Conakry Region Guinea "E"ETTC"C--SSLLEE--00121375 ETECT"--CS-LSEL--E0-2303226 " ETC-GIN-015 Kindia Guinea ET"C"-SLE-0116 Lofa Macenta Western " " ETC-SLE-009 ETC-GIN-017 Forecariah Guinea Area Rural " ETC-GIN-018 Conakry Guinea ETC-SLE-004 Nzerekore ETC-SLE-010 -

Guinea End of Project Report
Guinea Guinea Ebola Response and Recovery: Restoration and Improvement of Reproductive, Maternal, Newborn and Child Health Services (RHS) End of Project Report July 2015 - December 2016 Submitted to: United States Agency for International Development under Cooperative Agreement #AID-OAA-A-14-00028 Submitted by: Jhpiego Corporation The Maternal and Child Survival Program (MCSP) is a global United States Agency for International Development (USAID) Cooperative Agreement to introduce and support high-impact health interventions with a focus on 24 high-priority countries with the ultimate goal of ending preventable child and maternal deaths within a generation. The Program is focused on ensuring that all women, newborns and children most in need have equitable access to quality health care services to save lives. MCSP supports programming in maternal, newborn and child health, immunization, family planning and reproductive health, nutrition, health systems strengthening, water/sanitation/hygiene, malaria, prevention of mother-to-child transmission of HIV, and pediatric HIV care and treatment. Visit www.mcsprogram.org to learn more. This report is made possible by the generous support of the American people through USAID under the terms of the Cooperative Agreement AID-OAA-A-14-00028. The contents are the responsibility of MCSP and do not necessarily reflect the views of USAID or the United States Government. 2 Contents Acronyms and Abbreviations .........................................................................................................................................