Synthesis of New Polyether Ether Ketone Derivatives with Silver Binding Site and Coordination Compounds of Their Monomers with Different Silver Salts
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Surface Modifications of Poly(Ether Ether Ketone) Via Polymerization
materials Review Surface Modifications of Poly(Ether Ether Ketone) via Polymerization Methods—Current Status and Future Prospects Monika Flejszar and Paweł Chmielarz * Department of Physical Chemistry, Faculty of Chemistry, Rzeszow University of Technology, Al. Powsta´nców Warszawy 6, 35-959 Rzeszów, Poland; [email protected] * Correspondence: [email protected]; Tel.: +48-17-865-1809 Received: 24 January 2020; Accepted: 20 February 2020; Published: 23 February 2020 Abstract: Surface modification of poly(ether ether ketone) (PEEK) aimed at applying it as a bone implant material aroused the unflagging interest of the research community. In view of the development of implantology and the growing demand for new biomaterials, increasing biocompatibility and improving osseointegration are becoming the primary goals of PEEK surface modifications. The main aim of this review is to summarize the use of polymerization methods and various monomers applied for surface modification of PEEK to increase its bioactivity, which is a critical factor for successful applications of biomedical materials. In addition, the future directions of PEEK surface modifications are suggested, pointing to low-ppm surface-initiated atom transfer radical polymerization (SI-ATRP) as a method with unexplored capacity for flat surface modifications. Keywords: PEEK; surface modification; polymer brushes; ultraviolet (UV)-initiated graft polymerization; SI-ATRP 1. Introduction Currently, the production of bone implants is limited only to metal materials (stainless steel, cobalt–chromium, titanium). However, in the production of personalized bone implants, there is an alternative synthetic polymer named poly(ether ether ketone) (PEEK) [1–3]. The chemical structure of PEEK can be defined as an alternating combination of aryl rings through ketone and ether groups; therefore, it belongs to the family of polyaryletherketone polymers. -

Grignard Reaction: Synthesis of Triphenylmethanol
*NOTE: Grignard reactions are very moisture sensitive, so all the glassware in the reaction (excluding the work-up) should be dried in an oven with a temperature of > 100oC overnight. The following items require oven drying. They should be placed in a 150mL beaker, all labeled with a permanent marker. 1. 5mL conical vial (AKA: Distillation receiver). 2. Magnetic spin vane. 3. Claisen head. 4. Three Pasteur pipettes. 5. Two 1-dram vials (Caps EXCLUDED). 6. One 2-dram vial (Caps EXCLUDED). 7. Glass stirring rod 8. Adaptor (19/22.14/20) Grignard Reaction: Synthesis of Triphenylmethanol Pre-Lab: In the “equations” section, besides the main equations, also: 1) draw the equation for the production of the byproduct, Biphenyl. 2) what other byproduct might occur in the reaction? Why? In the “observation” section, draw data tables in the corresponding places, each with 2 columns -- one for “prediction” (by answering the following questions) and one for actual drops or observation. 1) How many drops of bromobenzene should you add? 2) How many drops of ether will you add to flask 2? 3) 100 µL is approximately how many drops? 4) What are the four signs of a chemical reaction? (Think back to Chem. 110) 5) How do the signs of a chemical reaction apply to this lab? The Grignard reaction is a useful synthetic procedure for forming new carbon- carbon bonds. This organometallic chemical reaction involves alkyl- or aryl-magnesium halides, known as Grignard 1 reagents. Grignard reagents are formed via the action of an alkyl or aryl halide on magnesium metal. -

Table 1. SOME NAMES and ABBREVIATIONS of Plastics and Elastomers
TABlE 1. SOME NAMES AND ABBREVIATIONS OF PlASTICS AND ElASTOMERS. Common name Abbreviation Acetal (homopolymer and copolymer) POM-H and POM-K Acrylate styrene acrylonitrile ASAorAAS Acrylate modified styrene acrylonitrile ASAorAAS Acrylic acid ester rubber ACM Acrylonitrile butadiene rubber or nitrile butadiene rubber NBR Acrylonitrile butadiene styrene ABS Acrylonitrile styrene/chlorinated polyethylene ACS Acrylonitrile methyl methacrylate AMMA Acrylonitrile styrene/EPR rubber or, acrylonitrile ethylene propylene styrene AES Alpha methyl styrene AMS Atactic polypropylene APPorPP-A Butadiene rubber or, cis-1,4-polybutadiene rubber or, polybutadiene rubber BR Butadiene styrene block copolymer BDS Butyl rubber IIR Bulk molding compound BMC Casein formaldehyde CF Cellulose acetate CA Cellulose acetate butyrate CAB Cellulose acetate propionate CAP Cellulose nitrate CN Chlorinated polyethylene CPEorCM Chlorinated polyvinyl chloride CPVC or, PVC-C Chloro-polyethylene or, chlorinated polyethylene. CM or CPE or, PE-C Chloroprene rubber or, polychloroprene rubber CR Chlorotrifluoroethylene ethylene copolymers ECfFE Cis-polyisoprene or, cis-1,4-polyisoprene IR Coumarone indene resins CIR Diallyl phthalate DAP Diallyl isophthalate DAIP Dough molding compound DMC Elastomeric alloy melt processable rubber EA-MPR Elastomeric alloy thermoplastic vulcanizate EA-TPV Epichlohydrin rubber CHR Epoxy or, epoxide EP Epoxy or, epoxide, with glass fiber EPGF Ethyl cellulose EC Ethylene acryic acid EAA Ethylene propylene diene monomer (an EPR terpolymer) EPDM -

13C NMR Study of Co-Contamination of Clays with Carbon Tetrachloride
Environ. Sci. Technol. 1998, 32, 350-357 13 sometimes make it the equal of solid acids like zeolites or C NMR Study of Co-Contamination silica-aluminas. Benesi (7-9) measured the Hammett acidity of Clays with Carbon Tetrachloride function H0 for a number of clays; these H0 values range from +1.5 to -8.2 (in comparison to H0 )-12 for 100% ) and Benzene sulfuric acid and H0 5 for pure acetic acid). Therefore, one can expect that certain chemical transformations might occur in/on clays that are similar to what are observed in zeolite TING TAO AND GARY E. MACIEL* systems. Thus, it is of interest to examine what happens Department of Chemistry, Colorado State University, when carbon tetrachloride and benzene are ªco-contami- Fort Collins, Colorado 80523 nantsº in a clay. This kind of information would be useful in a long-term view for understanding chemical transforma- tions of contaminants in soil at contaminated sites. Data on 13 these phenomena could also be useful for designing predic- Both solid-sample and liquid-sample C NMR experiments tive models and/or effective pollution remediation strategies. have been carried out to identify the species produced by Solid-state NMR results, based on 13C detection and line the reaction between carbon tetrachloride and benzene narrowing by magic angle spinning (MAS) and high-power when adsorbed on clays, kaolinite, and montmorillonite. Liquid- 1H decoupling (10), have been reported on a variety of organic sample 13C and 1H NMR spectra of perdeuteriobenzene soil components such as humic samples (11-13). Appar- extracts confirm the identities determined by solid-sample ently, there have been few NMR studies concerned directly 13C NMR and provide quantitative measures of the amounts with elucidating the interactions of organic compounds with of the products identifiedsbenzoic acid, benzophenone, and soil or its major components. -

On the Limits of Benzophenone As Cross-Linker for Surface-Attached Polymer Hydrogels
polymers Article On the Limits of Benzophenone as Cross-Linker for Surface-Attached Polymer Hydrogels Esther K. Riga †, Julia S. Saar †, Roman Erath, Michelle Hechenbichler and Karen Lienkamp * ID Freiburg Center für Interactive Materials and Bioinspired Technologies (FIT) and Department of Microsystems Engineering (IMTEK), Albert-Ludwigs-Universität, Georges-Köhler-Allee 105, 79110 Freiburg, Germany; [email protected] (E.K.R.); [email protected] (J.S.S.) * Correspondence: [email protected]; Tel.: +49-761-203-95090 † These authors contributed equally to this work. Received: 17 November 2017; Accepted: 4 December 2017; Published: 7 December 2017 Abstract: The synthesis of different photo-reactive poly(alkenyl norbornenes) and poly(oxonorbornenes) containing benzophenone (BP) via ring-opening metatheses polymerization (ROMP) is described. These polymers are UV irradiated to form well-defined surface-attached polymer networks and hydrogels. The relative propensity of the polymers to cross-link is evaluated by studying their gel content and its dependency on BP content, irradiation wavelength (254 or 365 nm) and energy dose applied (up to 11 J·cm−2). Analysis of the UV spectra of the polymer networks demonstrates that the poly(oxonorbornenes) show the expected BP-induced crosslinking behavior at 365 nm, although high irradiation energy doses and BP content are needed. However, these polymers undergo chain scission at 254 nm. The poly(alkenyl norbornenes), on the other hand, do not cross-link at 365 nm, whereas moderate to good cross-linking is observed at 254 nm. UV spectra demonstrate that the cross-linking at 254 nm is due to BP cross-linking combined with a [2 + 2] cylcoaddition of the alkenyl double bonds. -

Synthetic Polymeric Materials for Bone Replacement
Review Synthetic Polymeric Materials for Bone Replacement Mônica Rufino Senra and Maria de Fátima Vieira Marques * Technology Center, Bloco J, Instituto de Macromoleculas Professora Eloisa Mano, Universidade Federal do Rio de Janeiro, IMA-UFRJ, Av. Horacio Macedo, 2030, Rio de Janeiro RJ 21941-598, Brazil; [email protected] * Correspondence: [email protected] Received: 17 November 2020; Accepted: 18 December 2020; Published: 19 December 2020 Abstract: Some treatment options available to repair bone defects are the use of autogenous and allogeneic bone grafts. The drawback of the first one is the donor site’s limitation and the need for a second operation on the same patient. In the allograft method, the problems are associated with transmitted diseases and high susceptibility to rejection. As an alternative to biological grafts, polymers can be used in bone repair. Some polymers used in the orthopedic field are poly(methyl methacrylate), poly(ether-ether-ketone), and ultra-high molecular weight polyethylene (UHMWPE). UHMWPE has drawn much attention since it combines low friction coefficient and high wear and impact resistance. However, UHMWPE is a bioinert material, which means that it does not interact with the bone tissue. UHMWPE composites and nanocomposites with hydroxyapatite (HA) are widely studied in the literature to mitigate these issues. HA is the main component of the inorganic phase in the natural bone, and the addition of this bioactive filler to the polymeric matrix aims to mimic bone composition. This brief review discusses some polymers used in orthopedic applications, focusing on the UHMWPE/HA composites as a potential bone substitute. Keywords: poly(methyl methacrylate); poly(ether-ether-ketone); ultra-high molecular weight polyethylene; hydroxyapatite; orthopedic applications 1. -

Mechanical Properties Thermoplastic Laminates of Polycarbonate
Scienc ile e & Singh et al., J Textile Sci Eng 2018, 8:4 xt e E T n : g DOI 10.4172/2165-8064.1000366 f i o n l e a e n r r i n u g o Journal of Textile Science & Engineering J ISSN: 2165-8064 Research Article Article OpenOpen Access Access Mechanical Properties Thermoplastic Laminates of Polycarbonate- Polyetherimide Blend with Glass Fibers Singh JC1*, Srivastava M2, Singh RK2 and Yadaw SB3 1Indian Institute of Technology, New Delhi, India 2Central Institute of Plastics Engineering and Technology, Hajipur, India 3Defence Material & Stores Research & Development Establishment, Kanpur, India Abstract Thermoplastic Laminates of Polycarbonate (PC)/Polyetherimide (PEI) blend with glass fibers were prepared by solution blending of PC & PEI in different ratio of 100/0, 95/5, 90/10, 85/15 and 80/20 using Dichloromethane as solvent followed by applying the resulting blend solutions over the cut sheet size (30 × 25 cm) of woven mat glass fibers & dried overnight, then stacked up 12 flies of the impregnates and consolidated in a hot press at 300ºC and 3-4 ton (30 kg/cm2) pressure for a duration of one hour & cooled. The laminates of PC/PEI blends/woven mat glass fibers were studied for their mechanical properties viz: Tensile, Flexural, Impact and Inter Laminar Shear Strength and thermo-mechanical properties viz: TGA, DSC, DMA as well as examination of morphology by SEM. The mechanical properties of laminates of PC/PEI blends/woven mat glass fibers shows an increasing trend in Inter Laminar Shear Strength, Tensile and Flexural Strength upto 20% PEI while Impact Strength shows an increase upto 10% and decreases while the percentage of PEI increases. -
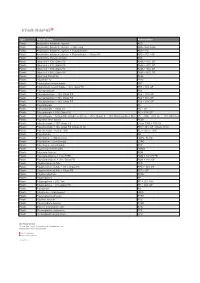
Type Material Name Abbreviation Plastic Acrylonitrile Butadiene
Type Material Name Abbreviation Plastic Acrylonitrile butadiene styrene ABS Plastic Acrylonitrile butadiene styrene - High-Temp ABS - high temp Plastic Acrylonitrile butadiene styrene + Polycarbonate ABS + PC Plastic Acrylonitrile butadiene styrene + Polycarbonate + Glass Fill ABS + PC + GF Plastic Acrylonitrile styrene acrylate ASA Plastic Nylon 6-6 + 10% Glass Fill PA66 + 10% GF Plastic Nylon 6-6 + 20% Glass Fill PA66 + 20% GF Plastic Nylon 6-6 + 30% Glass Fill PA66 + 30% GF Plastic Nylon 6-6 + 50% Glass Fill PA66 + 50% GF Plastic Nylon 6-6 Polyamide PA66 Plastic Polyamide 12 PA12 Plastic Polybutylene terephthalate PBT Plastic Polybutylene terephthalate + 30% Glass Fill PBT+ 30% GF Plastic Polycaprolactam PA6 Plastic Polycaprolactam + 20% Glass Fill PA6 + 20% GF Plastic Polycaprolactam + 30% Glass Fill PA6 + 30% GF Plastic Polycaprolactam + 50% Glass Fill PA6 + 50% GF Plastic Polycarbonate PC Plastic Polycarbonate + Glass Fill PC + GF Plastic Polycarbonate + 10% Glass Fill PC + 10% GF Plastic Polycarbonate + Acrylonitrile butadiene styrene + 20% Glass Fill + 10% Stainless Steel fiber PC + ABS + 20% GF + 10% SS Fiber Plastic Polyether ether ketone PEEK Plastic Polyetherimide + 30% Glass Fill Ultem 1000 + 30% GF Plastic Polyetherimide + 40% Glass Fill (Ultem 2410) PEI + 40% GF (Ultem 2410) Plastic Polyetherimide + Ultem 1000 PEI + Ultem 1000 Plastic Polyethylene PE Plastic Polyethylene - High-Density HDPE, PEHD Plastic Polyethylene - Low-Density LDPE Plastic Polyethylene terephthalate PET Plastic Polymethyl methacrylate PMMA Plastic Polyoxymethylene -

Impact Strength of 3D Printed Polyether-Ether-Ketone (PEEK)
Solid Freeform Fabrication 2019: Proceedings of the 30th Annual International Solid Freeform Fabrication Symposium – An Additive Manufacturing Conference Impact Strength of 3D Printed Polyether-ether-ketone (PEEK) Haijun Gong1,*, Xiaodong Xing2, Jan Nel1 1Department of Manufacturing Engineering, Georgia Southern University, Statesboro GA, USA 2College of Mechanical and Electrical Engineering, Harbin Engineering University, Harbin, China *Corresponding author ([email protected]) Abstract Polyether-ether-ketone (PEEK) is a high-performance thermoplastic with high heat-, high chemical-, high water-, and high wear-resistance. Its strength and durability also make it highly accepted for a range of industrial applications. 3D printing of PEEK filaments offers a unique quality and flexibility in making PEEK parts for low-volume production or special designs. This study investigates the impact strength of 3D printed PEEK materials. The specimens are fabricated using a fused deposition modeling (FDM) based 3D printer and tested by a pendulum impact tester in compliance with ASTM standard. The testing result is discussed with respect to the processing parameters and the annealing treatment. Impact strength comparison of PEEK materials manufactured by 3D printing and by conventional production is also conducted. Instruction Polyether-ether-ketone (PEEK) is a semi-crystalline polymer, which was developed in 1978. The PEEK is widely used as a high-performance thermoplastic due to its high heat-, high chemical-, high water-, high wear-resistance. Especially, PEEK and its varieties have been adopted for different biomedical treatment such as dental implants, orthopedics and maxillofacial surgery [1,2]. Its strength and durability also make it highly accepted for a range of industrial applications. For example, many automotive applications require to work at a temperature more than 120 °C. -

Ether Ether Ketone): Monitoring the Meta-Fluorine Displacement in 3,5,4’- Trifluorobenzophenone
Wright State University CORE Scholar Browse all Theses and Dissertations Theses and Dissertations 2017 Toward The Synthesis of Functionalized Poly (Ether Ether Ketone): Monitoring the meta-Fluorine Displacement in 3,5,4’- trifluorobenzophenone Giovanni Covarrubias Wright State University Follow this and additional works at: https://corescholar.libraries.wright.edu/etd_all Part of the Chemistry Commons Repository Citation Covarrubias, Giovanni, "Toward The Synthesis of Functionalized Poly (Ether Ether Ketone): Monitoring the meta-Fluorine Displacement in 3,5,4’-trifluorobenzophenone" (2017). Browse all Theses and Dissertations. 1725. https://corescholar.libraries.wright.edu/etd_all/1725 This Thesis is brought to you for free and open access by the Theses and Dissertations at CORE Scholar. It has been accepted for inclusion in Browse all Theses and Dissertations by an authorized administrator of CORE Scholar. For more information, please contact [email protected]. TOWARD THE SYNTHESIS OF FUNCTIONALIZED, SEMI-CRYSTALLINE POLY (ETHER ETHER KETONE): MONITORING THE META-FLUORINE DISPLACEMENT IN 3,5,4’-TRIFLUOROBENZOPHENONE A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science By: GIOVANNI COVARRUBIAS B.S. Loras College, 2013 2017 Wright State University WRIGHT STATE UNIVERSITY GRADUATE SCHOOL May 18th, 2017 I HEREBY RECOMMEND THAT THE THESIS PREPARED UNDER MY SUPERVISION BY Giovanni Covarrubias ENTITLED Toward The Synthesis of Functionalized Poly (Ether Ether Ketone)s: Monitoring the meta-Fluorine Displacement in 3,5,4’-trifluorobenzophenone BE ACCEPTED IN PARTIAL FULLFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Master of Science. ____________________________ Eric Fossum, Ph. D. Thesis Advisor ____________________________ David Grossie, Ph. D. Chair, Department of Chemistry Committee on Final Examination _____________________________ Eric Fossum, Ph. -

Material Safety Data Sheet HCS Risk Phrases HCS CLASS: Corrosive Liquid
Material Safety Data Sheet HCS Risk Phrases HCS CLASS: Corrosive liquid. HCS CLASS: Combustible liquid IIIA having a flash point between 60.0°C (140°F) and 93.3°C (200°F) Section I. Chemical Product and Company Identification Common Name/ Benzoyl Chloride In Case of Trade Name Emergency In the continental U.S.A. call CHEMTREC 800-424-9300 (24 hours) Outside the continental U.S.A. call CHEMTREC 703-527-3887 (24 hours) Supplier Velsicol Chemical Corporation Manufacturer Velsicol Chemical Corporation 10400 W. Higgins Road 10400 W. Higgins Road Rosemont, IL 60018 U.S.A. Rosemont, IL 60018 U.S.A. Phone: 847-298-9000 Phone: 847-298-9000 Fax: 847-298-9015 Fax: 847-298-9015 Synonym Benzenecarbonyl Chloride Material Uses Agricultural Industry: Chloramber Chemical Name Benzoyl Chloride (herbicide). Intermediate for pesticides. Chemical Family Acyl Halide Industrial Applications: Acylation. Chemical C7 H5 CL 0 Polymerization initiator of benzophenone. Formula Intermediate for stabilizers. Acetic anhydride production. Textile Industry: Cellulosic yarn treatment agent. Fastness improver. Other Non-Specified Industry: Organic analysis. Dyes. Perfumes. Section II. Composition and Information on Ingredients Name CAS# % by Weight TLV/PEL OSHA Hazardous Ingredients 1) Benzoyl Chloride 98-88-4 >99.5 STEL: 2.8 (mg/m3) from Yes ACGIH (TLV) 2) Benzotrichloride 98-07-7 <0.02 STEL: 0.1 ppm from Yes ACGIH; Skin designation. 3) Benzyl Alcohol 100-44-7 <0.01 TWA: 1 (ppm) from Yes ACGIH (TLV) Section III. Hazards Identification Emergency Overview Clear. Liquid. Pungent. Acrid odor. DANGER! CAUSES SEVERE EYE, SKIN AND RESPIRATORY TRACT BURNS. MAY CAUSE BLINDNESS. -

Synthesis and Process Optimization of Electrospun PEEK-Sulfonated Nanofibers by Response Surface Methodology
Materials 2015, 8, 4096-4117; doi:10.3390/ma8074096 OPEN ACCESS materials ISSN 1996-1944 www.mdpi.com/journal/materials Article Synthesis and Process Optimization of Electrospun PEEK-Sulfonated Nanofibers by Response Surface Methodology Carlo Boaretti, Martina Roso, Alessandra Lorenzetti and Michele Modesti * Department of Industrial Engineering, University of Padova, via Marzolo 9, 35131 Padova, Italy; E-Mails: [email protected] (C.B.); [email protected] (M.R.); [email protected] (A.L.) * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +39-049-827-5541; Fax: +39-049-827-5555. Academic Editor: Maryam Tabrizian Received: 22 May 2015 / Accepted: 30 June 2015 / Published: 7 July 2015 Abstract: In this study electrospun nanofibers of partially sulfonated polyether ether ketone have been produced as a preliminary step for a possible development of composite proton exchange membranes for fuel cells. Response surface methodology has been employed for the modelling and optimization of the electrospinning process, using a Box-Behnken design. The investigation, based on a second order polynomial model, has been focused on the analysis of the effect of both process (voltage, tip-to-collector distance, flow rate) and material (sulfonation degree) variables on the mean fiber diameter. The final model has been verified by a series of statistical tests on the residuals and validated by a comparison procedure of samples at different sulfonation degrees, realized according to optimized conditions, for the production of homogeneous thin nanofibers. Keywords: electrospinning; response surface methodology; sulfonated polyether ether ketone 1. Introduction Nanofibers are an interesting and versatile class of one-dimensional nanomaterials, with diameters ranging from tenths to hundreds of nanometers, which have been recognized as promising due to their outstanding properties in terms of high porosity, excellent pore interconnectivity, small diameters and high surface-to-volume ratio.