Induced Changes in Gene Expression in Human CNS Tumors As Determined by U95av2 Chip Oligoarray Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Examples of Amino Acid Changes in Notothenioids Methods To
Supplementary Information S1 Candidate genes: examples of amino acid changes in notothenioids Methods To evaluate notothenioid amino acid changes in a cross‐section of genes, six candidates from a range of functional categories of interest were chosen for a more detailed examination. The selected genes comprised small, well‐characterised genes present in multiple fish species (mainly teleosts, but in some cases these analyses were extended to the Actinopterygii when sequences from the spotted gar (Lepisosteus oculatus) were available) and available in at least two notothenioids. The amino acid composition of a series of six candidate genes were further analysed in depth at the amino acid sequence level. These genes were superoxide dismutase 1 (SOD1), neuroglobin, dihydrofolate reductase (both paralogues), p53 and calmodulin. Clustal X alignments were constructed from orthologues identified in the four Notothenioid transcriptomes along with other fish orthologues extracted from either SwissProt (Bateman et al., 2017) or Ensembl release 91 (Aken et al., 2017). Alignment data were visualised and annotated in BoxShade version 3.21 (https://embnet.vital‐ it.ch/software/BOX_form.html). Notothenioid‐specific changes were noted and the type of substitution in terms of amino acid properties was noted. Results A range of amino acid changes were revealed, from virtually complete conservation between the notothenioids and other fish (calmodulin), to one amino acid substitution present in neuroglobin and SOD1, to more extensive changes in dihydrofolate reductase, dihydrofolate reductase‐like and p53. In the case of the latter three genes, notothenioid‐specific changes were at positions that were often highly variable in temperate fish species with up to six different amino acid substitutions and did not result in a definable pattern of directional substitution. -

MAP2K3 (Human) Recombinant Protein (Q01)
MAP2K3 (Human) Recombinant phosphorylates and thus activates MAPK14/p38-MAPK. Protein (Q01) This kinase can be activated by insulin, and is necessary for the expression of glucose transporter. Expression of Catalog Number: H00005606-Q01 RAS oncogene is found to result in the accumulation of the active form of this kinase, which thus leads to the Regulation Status: For research use only (RUO) constitutive activation of MAPK14, and confers oncogenic transformation of primary cells. The inhibition Product Description: Human MAP2K3 partial ORF ( of this kinase is involved in the pathogenesis of Yersina AAH32478, 1 a.a. - 100 a.a.) recombinant protein with pseudotuberculosis. Multiple alternatively spliced GST-tag at N-terminal. transcript variants that encode distinct isoforms have been reported for this gene. [provided by RefSeq] Sequence: MESPASSQPASMPQSKGKSKRKKDLRISCMSKPPAP NPTPPRNLDSRTFITIGDRNFEVEADDLVTISELGRGAY GVVEKVRHAQSGTIMAVKRIRATVN Host: Wheat Germ (in vitro) Theoretical MW (kDa): 36.63 Applications: AP, Array, ELISA, WB-Re (See our web site product page for detailed applications information) Protocols: See our web site at http://www.abnova.com/support/protocols.asp or product page for detailed protocols Preparation Method: in vitro wheat germ expression system Purification: Glutathione Sepharose 4 Fast Flow Storage Buffer: 50 mM Tris-HCI, 10 mM reduced Glutathione, pH=8.0 in the elution buffer. Storage Instruction: Store at -80°C. Aliquot to avoid repeated freezing and thawing. Entrez GeneID: 5606 Gene Symbol: MAP2K3 Gene Alias: MAPKK3, MEK3, MKK3, PRKMK3 Gene Summary: The protein encoded by this gene is a dual specificity protein kinase that belongs to the MAP kinase kinase family. This kinase is activated by mitogenic and environmental stress, and participates in the MAP kinase-mediated signaling cascade. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -

MAPK Kinase 3 Is a Tumor Suppressor with Reduced Copy Number in Breast Cancer
Author Manuscript Published OnlineFirst on November 14, 2013; DOI: 10.1158/0008-5472.CAN-13-1310 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. MAPK kinase 3 is a tumor suppressor with reduced copy number in breast cancer Adam J. MacNeil1,2,3, Shun-Chang Jiao4, Lori A. McEachern5, Yong Jun Yang6, Amanda Dennis1,2, Haiming Yu4, Zhaolin Xu3,7, Jean S. Marshall1,3, and Tong-Jun Lin1,2,3,7 1 Department of Microbiology & Immunology, Dalhousie University, Halifax, Nova Scotia, B3K 6R8, Canada. 2 Department of Pediatrics, Dalhousie University, Halifax, Nova Scotia, B3K 6R8, Canada. 3 Beatrice Hunter Cancer Research Institute, Suite 2L-A2, Tupper Link, 5850 College Street, PO Box 15000, Halifax, Nova Scotia, B3H 4R2, Canada. 4 Department of Medical Oncology, General Hospital of the People's Liberation Army, Beijing, 100853, China. 5 Department of Physiology & Biophysics, Dalhousie University, Halifax, Nova Scotia, B3K 6R8, Canada. 6 Institute of Zoonosis, College of Animal Sciences and Veterinary Medicine, Jilin University, Changchun, Jilin, 130062, China. 7 Department of Pathology, Dalhousie University, Halifax, Nova Scotia, B3K 6R8, Canada. Correspondence: Shun-Chang Jiao, email:[email protected]; Department of Medical Oncology, General Hospital of the People's Liberation Army, Beijing, 100853, China. Tong-Jun Lin, email: [email protected]; phone 902-470-8834; IWK Health Centre, Department of Pediatrics, Dalhousie University, 5850/5980 University Ave, Halifax, Nova Scotia, B3K 6R8, Canada. The authors have declared that no conflict of interest exists. Downloaded from cancerres.aacrjournals.org on September 29, 2021. © 2013 American Association for Cancer Research. -

Integrated Molecular Characterisation of the MAPK Pathways in Human
ARTICLE https://doi.org/10.1038/s42003-020-01552-6 OPEN Integrated molecular characterisation of the MAPK pathways in human cancers reveals pharmacologically vulnerable mutations and gene dependencies 1234567890():,; ✉ Musalula Sinkala 1 , Panji Nkhoma 2, Nicola Mulder 1 & Darren Patrick Martin1 The mitogen-activated protein kinase (MAPK) pathways are crucial regulators of the cellular processes that fuel the malignant transformation of normal cells. The molecular aberrations which lead to cancer involve mutations in, and transcription variations of, various MAPK pathway genes. Here, we examine the genome sequences of 40,848 patient-derived tumours representing 101 distinct human cancers to identify cancer-associated mutations in MAPK signalling pathway genes. We show that patients with tumours that have mutations within genes of the ERK-1/2 pathway, the p38 pathways, or multiple MAPK pathway modules, tend to have worse disease outcomes than patients with tumours that have no mutations within the MAPK pathways genes. Furthermore, by integrating information extracted from various large-scale molecular datasets, we expose the relationship between the fitness of cancer cells after CRISPR mediated gene knockout of MAPK pathway genes, and their dose-responses to MAPK pathway inhibitors. Besides providing new insights into MAPK pathways, we unearth vulnerabilities in specific pathway genes that are reflected in the re sponses of cancer cells to MAPK targeting drugs: a revelation with great potential for guiding the development of innovative therapies. -

Temperature Regulates NF-Κb Dynamics and Function Through Timing of A20 Transcription
Temperature regulates NF-κB dynamics and function through timing of A20 transcription C. V. Harpera,1, D. J. Woodcockb,c,1, C. Lama, M. Garcia-Albornozd, A. Adamsona, L. Ashalla, W. Rowea, P. Downtona, L. Schmidta, S. Weste, D. G. Spillera, D. A. Randb,c,2, and M. R. H. Whitea,2 aSystems Microscopy Centre, Division of Molecular and Cellular Function, School of Biology, Faculty of Biology, Medicine and Health, Manchester Academic Health Sciences Centre, University of Manchester, M13 9PT Manchester, United Kingdom; bMathematics Institute, University of Warwick, CV4 7AL Coventry, United Kingdom; cZeeman Institute for Systems Biology and Infectious Epidemiology Research, University of Warwick, CV4 7AL Coventry, United Kingdom; dDivision of Evolution and Genomic Sciences, School of Biology, Faculty of Biology, Medicine and Health, Manchester Academic Health Sciences Centre, University of Manchester, M13 9PT Manchester, United Kingdom; and eInstitute of Integrative Biology, University of Liverpool, L69 7ZB United Kingdom Edited by Ronald N. Germain, National Institutes of Health, Bethesda, MD, and approved April 27, 2018 (received for review March 7, 2018) NF-κB signaling plays a pivotal role in control of the inflammatory by measuring both the oscillatory dynamics and the downstream gene response. We investigated how the dynamics and function of NF-κB expression response. were affected by temperature within the mammalian physiological range (34 °C to 40 °C). An increase in temperature led to an increase Results in NF-κB nuclear/cytoplasmic oscillation frequency following Tumor To investigate the effect of temperature on NF-κB dynamics, we Necrosis Factor alpha (TNFα) stimulation. Mathematical modeling used time-lapse live-cell microscopy to study cytoplasmic to nu- suggested that this temperature sensitivity might be due to an clear oscillations of NF-κB across the physiological temper- A20-dependent mechanism, and A20 silencing removed the sensi- ature range, 34 °C to 40 °C. -

Delineation of Key Regulatory Elements Identifies Points Of
DELINEATION OF KEY REGULATORY ELEMENTS IDENTIFIES POINTS OF VULNERABILITY IN THE MITOGEN-ACTIVATED SIGNALING NETWORK SUPPLEMENTARY MATERIALS List of contents Supplementary Figures with legends 1. Figure S1: Distribution of primary siRNA screen data, and standardization of assay procedure. 2. Figure S2: Scatter plot of screen data. 3. Figure S3: Functional relevance of the identified targets and Calculation of residence time from PDT and cell cycle distribution. 4. Figure S4: FACS profiles for ABL1 and AKT1. Table for data in Figure 5B. 5. Figure S5: Venn diagram showing the results of the comparative analysis of other screen results 6. Figure S6: Dose response profiles for the AKT1 + ABL1 inhibitor combination for CH1, list of the 14 cell lines and their description, effect of ABL1+AKT1 inhibitor combination on increase in apoptotic cells and G1 arrest in 14 cell lines, effects of CHEK1 inhibitor on combination C1,C2 on 4 cell lines. Supplementary Tables 1. Table S1: siRNA screen results for targeted kinases and phosphatases. 2. Table S2: Gene expression status of the validated hits. 3. Table S3: Role played by identified RNAi hits in regulation of cell cycle, the effect on PDTs along with phase-specific RTs. 4. Table S4: List of molecules classified as cell cycle targets. 5. Table S5: High confidence network used for graph theory analysis. 6. Table S6: Occurrences of nodes in shortest path networks. 7. Table S7: Network file used as SNAVI background. 8. Table S8: Classification of nodes present in modules according to specificity. Legends for tables Supplementary Experimental Procedures References Figure S1 A 450 400 G1 S 350 G2 300 250 200 150 100 50 Distribution of molecules Distribution 0 -6-4-20246 Z-score 350 200 400 G1 S 300 G2 150 300 250 200 100 200 150 100 50 100 Distribution of molecules 50 0 0 0 -4 -2 0 2 4 -4-20246 -4-20246 Z-score B PLK1 GAPDH PLCg BTK PLCg CDC2A PLCg CHEK1 PLCg MET Distribution profiles of complete primary screen and western blots showing knockdown efficiency. -
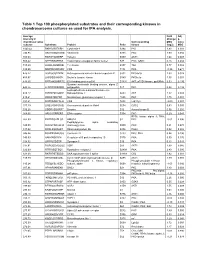
Table 1 Top 100 Phosphorylated Substrates and Their Corresponding Kinases in Chondrosarcoma Cultures As Used for IPA Analysis
Table 1 Top 100 phosphorylated substrates and their corresponding kinases in chondrosarcoma cultures as used for IPA analysis. Average Fold Adj intensity in Change p- chondrosarcoma Corresponding MSC value cultures Substrate Protein Psite kinase (log2) MSC 1043.42 RKKKVSSTKRH Cytohesin-1 S394 PKC 1.83 0.001 746.95 RKGYRSQRGHS Vitronectin S381 PKC 1.00 0.056 709.03 RARSTSLNERP Tuberin S939 AKT1 1.64 0.008 559.42 SPPRSSLRRSS Transcription elongation factor A-like1 S37 PKC; GSK3 0.18 0.684 515.29 LRRSLSRSMSQ Telethonin S157 Titin 0.77 0.082 510.00 MQPDNSSDSDY CD5 T434 PKA -0.35 0.671 476.27 GGRGGSRARNL Heterogeneous nuclear ribonucleoprotein K S302 PKCdelta 1.03 0.028 455.97 LKPGSSHRKTK Bruton's tyrosine kinase S180 PKCbeta 1.55 0.001 444.65 RRRMASMQRTG E1A binding protein p300 S1834 AKT; p70S6 kinase; pp90Rsk 0.53 0.195 Guanine nucleotide binding protein, alpha Z 440.26 HLRSESQRQRR polypeptide S27 PKC 0.88 0.199 6-phosphofructo-2-kinase/fructose-2,6- 424.12 RPRNYSVGSRP biphosphatase 2 S483 AKT 1.32 0.003 419.61 KKKIATRKPRF Metabotropic glutamate receptor 1 T695 PKC 1.75 0.001 391.21 DNSSDSDYDLH CD5 T453 Lck; Fyn -2.09 0.001 377.39 LRQLRSPRRAQ Ras associated protein Rab4 S204 CDC2 0.63 0.091 376.28 SSQRVSSYRRT Desmin S12 Aurora kinase B 0.56 0.255 369.05 ARIGGSRRERS EP4 receptor S354 PKC 0.29 0.543 RPS6 kinase alpha 3; PKA; 367.99 EPKRRSARLSA HMG14 S7 PKC -0.01 0.996 Peptidylglycine alpha amidating 349.08 SRKGYSRKGFD monooxygenase S930 PKC 0.21 0.678 347.92 RRRLSSLRAST Ribosomal protein S6 S236 PAK2 0.02 0.985 346.84 RSNPPSRKGSG Connexin -

Short-Hairpin RNA Library: Identification of Therapeutic Partners for Gefitinib-Resistant Non-Small Cell Lung Cancer
www.impactjournals.com/oncotarget/ Oncotarget, Vol. 6, No.2 Short-hairpin RNA library: identification of therapeutic partners for gefitinib-resistant non-small cell lung cancer Makoto Sudo1, Seiichi Mori2, Vikas Madan1, Henry Yang1, Geraldine Leong1 and H. Phillip Koeffler1,3,4 1 Cancer Science Institute of Singapore, NUS, Singapore 2 Division of Cancer Genomics, The Cancer Institute of Japanese Foundation for Cancer Research, Tokyo, Japan 3 Department of Hematology and Oncology, Cedars-Sinai Medical Center, Los Angeles, CA, USA 4 National University Cancer Institute, National University Hospital, Singapore Correspondence to: Makoto Sudo, email: [email protected] Keywords: combination chemotherapy, dasatinib, gefitinib, PRKCSH, short-hairpin RNA library screening, thioridazine Received: August 19, 2014 Accepted: November 24, 2014 Published: November 25, 2014 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Somatic mutations of the epidermal growth factor receptor often cause resistance to therapy with tyrosine kinase inhibitor in non-small cell lung cancer (NSCLC). In this study, we aimed to identify partner drugs and pathways that can induce cell death in combination with gefitinib in NSCLC cells. We undertook a genome-wide RNAi screen to identify synthetic lethality with gefitinib in tyrosine kinase inhibitor resistant cells. The screening data were utilized in different approaches. Firstly, we identifiedPRKCSH as a candidate gene, silencing of which induces apoptosis of NSCLC cells treated with gefitinib. Next, in an in silico gene signature pathway analysis of shRNA library data, a strong correlation of genes involved in the CD27 signaling cascade was observed. -

Knockdown of Mitogen-Activated Protein Kinase Kinase 3 Negatively Regulates Hepatitis a Virus Replication
International Journal of Molecular Sciences Article Knockdown of Mitogen-Activated Protein Kinase Kinase 3 Negatively Regulates Hepatitis A Virus Replication Tatsuo Kanda 1,* , Reina Sasaki-Tanaka 1, Ryota Masuzaki 1 , Naoki Matsumoto 1, Hiroaki Okamoto 2 and Mitsuhiko Moriyama 1 1 Division of Gastroenterology and Hepatology, Department of Medicine, Nihon University School of Medicine, 30-1 Oyaguchi-kamicho, Itabashi-ku, Tokyo 173-8610, Japan; [email protected] (R.S.-T.); [email protected] (R.M.); [email protected] (N.M.); [email protected] (M.M.) 2 Division of Virology, Department of Infection and Immunity, Jichi Medical University School of Medicine, Shimotsuke, Tochigi 329-0498, Japan; [email protected] * Correspondence: [email protected]; Tel.: +81-3-3972-8111 Abstract: Zinc chloride is known to be effective in combatting hepatitis A virus (HAV) infection, and zinc ions seem to be especially involved in Toll-like receptor (TLR) signaling pathways. In the present study, we examined this involvement in human hepatoma cell lines using a human TLR signaling target RT-PCR array. We also observed that zinc chloride inhibited mitogen-activated protein kinase kinase 3 (MAP2K3) expression, which could downregulate HAV replication in human hepatocytes. It is possible that zinc chloride may inhibit HAV replication in association with its inhibition of MAP2K3. In that regard, this study set out to determine whether MAP2K3 could be considered a modulating factor in the development of the HAV pathogen-associated molecular pattern (PAMP) Citation: Kanda, T.; Sasaki-Tanaka, and its triggering of interferon-β production. -

A Novel Glycogen Synthase Kinase-3 Inhibitor Optimized for Acute
Published OnlineFirst May 9, 2016; DOI: 10.1158/1535-7163.MCT-15-0566 Small Molecule Therapeutics Molecular Cancer Therapeutics A Novel Glycogen Synthase Kinase-3 Inhibitor Optimized for Acute Myeloid Leukemia Differentiation Activity Sophia Hu1, Masumi Ueda2, Lindsay Stetson1, James Ignatz-Hoover1, Stephen Moreton1, Amit Chakrabarti3, Zhiqiang Xia3, Goutam Karan3, Marcos de Lima2, Mukesh K. Agrawal3,4, and David N. Wald1,3,5 Abstract Standard therapies used for the treatment of acute myeloid describe the discovery of a novel GSK3 inhibitor, GS87. GS87 leukemia (AML) are cytotoxic agents that target rapidly prolifer- was discovered in efforts to optimize GSK3 inhibition for AML ating cells. Unfortunately, this therapeutic approach has differentiation activity. Despite GS87's dramatic ability to induce limited efficacy and significant toxicity and the majority of AML AML differentiation, kinase profiling reveals its high specificity patients still die of their disease. In contrast to the poor prognosis in targeting GSK3 as compared with other kinases. GS87 demon- of most AML patients, most individuals with a rare subtype of strates high efficacy in a mouse AML model system and unlike AML, acute promyelocytic leukemia, can be cured by differenti- current AML therapeutics, exhibits little effect on normal bone ation therapy using regimens containing all-trans retinoic acid. marrow cells. GS87 induces potent differentiation by more GSK3 has been previously identified as a therapeutic target in effectively activating GSK3-dependent signaling components AML where its inhibition can lead to the differentiation and including MAPK signaling as compared with other GSK3 growth arrest of leukemic cells. Unfortunately, existing GSK3 inhibitors. GS87 is a novel GSK3 inhibitor with therapeutic inhibitors lead to suboptimal differentiation activity making potential as a differentiation agent for non-promyelocytic AML.