Knockdown of Mitogen-Activated Protein Kinase Kinase 3 Negatively Regulates Hepatitis a Virus Replication
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MAP2K3 (Human) Recombinant Protein (Q01)
MAP2K3 (Human) Recombinant phosphorylates and thus activates MAPK14/p38-MAPK. Protein (Q01) This kinase can be activated by insulin, and is necessary for the expression of glucose transporter. Expression of Catalog Number: H00005606-Q01 RAS oncogene is found to result in the accumulation of the active form of this kinase, which thus leads to the Regulation Status: For research use only (RUO) constitutive activation of MAPK14, and confers oncogenic transformation of primary cells. The inhibition Product Description: Human MAP2K3 partial ORF ( of this kinase is involved in the pathogenesis of Yersina AAH32478, 1 a.a. - 100 a.a.) recombinant protein with pseudotuberculosis. Multiple alternatively spliced GST-tag at N-terminal. transcript variants that encode distinct isoforms have been reported for this gene. [provided by RefSeq] Sequence: MESPASSQPASMPQSKGKSKRKKDLRISCMSKPPAP NPTPPRNLDSRTFITIGDRNFEVEADDLVTISELGRGAY GVVEKVRHAQSGTIMAVKRIRATVN Host: Wheat Germ (in vitro) Theoretical MW (kDa): 36.63 Applications: AP, Array, ELISA, WB-Re (See our web site product page for detailed applications information) Protocols: See our web site at http://www.abnova.com/support/protocols.asp or product page for detailed protocols Preparation Method: in vitro wheat germ expression system Purification: Glutathione Sepharose 4 Fast Flow Storage Buffer: 50 mM Tris-HCI, 10 mM reduced Glutathione, pH=8.0 in the elution buffer. Storage Instruction: Store at -80°C. Aliquot to avoid repeated freezing and thawing. Entrez GeneID: 5606 Gene Symbol: MAP2K3 Gene Alias: MAPKK3, MEK3, MKK3, PRKMK3 Gene Summary: The protein encoded by this gene is a dual specificity protein kinase that belongs to the MAP kinase kinase family. This kinase is activated by mitogenic and environmental stress, and participates in the MAP kinase-mediated signaling cascade. -

Galectin-4 Interaction with CD14 Triggers the Differentiation of Monocytes Into Macrophage-Like Cells Via the MAPK Signaling Pathway
Immune Netw. 2019 Jun;19(3):e17 https://doi.org/10.4110/in.2019.19.e17 pISSN 1598-2629·eISSN 2092-6685 Original Article Galectin-4 Interaction with CD14 Triggers the Differentiation of Monocytes into Macrophage-like Cells via the MAPK Signaling Pathway So-Hee Hong 1,2,3,4,5, Jun-Seop Shin 1,2,3,5, Hyunwoo Chung 1,2,4,5, Chung-Gyu Park 1,2,3,4,5,* 1Xenotransplantation Research Center, Seoul National University College of Medicine, Seoul 03080, Korea 2Institute of Endemic Diseases, Seoul National University College of Medicine, Seoul 03080, Korea 3Cancer Research Institute, Seoul National University College of Medicine, Seoul 03080, Korea 4Department of Biomedical Sciences, Seoul National University College of Medicine, Seoul 03080, Korea 5Department of Microbiology and Immunology, Seoul National University College of Medicine, Seoul 03080, Korea Received: Jan 28, 2019 ABSTRACT Revised: May 13, 2019 Accepted: May 19, 2019 Galectin-4 (Gal-4) is a β-galactoside-binding protein mostly expressed in the gastrointestinal *Correspondence to tract of animals. Although intensive functional studies have been done for other galectin Chung-Gyu Park isoforms, the immunoregulatory function of Gal-4 still remains ambiguous. Here, we Department of Microbiology and Immunology, Seoul National University College of Medicine, demonstrated that Gal-4 could bind to CD14 on monocytes and induce their differentiation 103 Daehak-ro, Jongno-gu, Seoul 03080, into macrophage-like cells through the MAPK signaling pathway. Gal-4 induced the phenotypic Korea. changes on monocytes by altering the expression of various surface molecules, and induced E-mail: [email protected] functional changes such as increased cytokine production and matrix metalloproteinase expression and reduced phagocytic capacity. -

Targeting the Mitogen-Activated Protein Kinase Pathway: Physiological Feedback and Drug Response
Published OnlineFirst May 14, 2010; DOI: 10.1158/1078-0432.CCR-09-3064 Molecular Pathways Clinical Cancer Research Targeting the Mitogen-Activated Protein Kinase Pathway: Physiological Feedback and Drug Response Christine A. Pratilas1,2 and David B. Solit3,4 Abstract Mitogen-activated protein kinase (MAPK) pathway activation is a frequent event in human cancer and is often the result of activating mutations in the BRAF and RAS oncogenes. Targeted inhibitors of BRAF and its downstream effectors are in various stages of preclinical and clinical development. These agents offer the possibility of greater efficacy and less toxicity than current therapies for tumors driven by oncogenic mutations in the MAPK pathway. Early clinical results with the BRAF-selective in- hibitor PLX4032 suggest that this strategy will prove successful in a select group of patients whose tu- mors are driven by V600E BRAF. Relief of physiologic feedback upon pathway inhibition may, however, attenuate drug response and contribute to the development of acquired resistance. An im- proved understanding of the adaptive response of cancer cells to MAPK pathway inhibition may thus aid in the identification of those patients most likely to respond to targeted pathway inhibitors and provide a rational basis for tailored combination strategies. Clin Cancer Res; 16(13); 3329–34. ©2010 AACR. Background these genetic alterations activate common downstream effectors of transformation. One of the central regulators of growth factor-induced Activating BRAF mutations are found clustered within cell proliferation and survival in both normal and cancer the P-loop (exon 11) and activation segment (exon 15) cells is the RAS protein. -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -

Fine Mapping and Expression of Candidate Genes Within the Chromosome 10 QTL Region of the High and Low Alcohol Drinking Rats
View metadata, citation and similar papers at core.ac.uk brought to you by CORE HHS Public Access provided by IUPUIScholarWorks Author manuscript Author ManuscriptAuthor Manuscript Author Alcohol Manuscript Author . Author manuscript; Manuscript Author available in PMC 2017 July 18. Published in final edited form as: Alcohol. 2010 September ; 44(6): 477–485. doi:10.1016/j.alcohol.2010.06.004. Fine mapping and expression of candidate genes within the Chromosome 10 QTL region of the High and Low Alcohol Drinking Rats Paula J. Bice1, Tiebing Liang1, Lili Zhang1, Tamara J. Graves1, Lucinda G. Carr1, Dongbing Lai2, Mark W. Kimpel3, and Tatiana Foroud2 1Department of Medicine, Indiana University School of Medicine, Indianapolis, Indiana 46202, USA 2Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN 46202, USA 3Department of Psychiatry, Indiana University School of Medicine, Indianapolis, IN 46202, USA Abstract The high and low alcohol-drinking (HAD and LAD) rats were selectively bred for differences in alcohol intake. The HAD/LAD rats originated from the N/Nih heterogeneous stock (HS) developed from intercrossing 8 inbred rat strains. The HAD × LAD F2 were genotyped, and a powerful analytical approach, utilizing ancestral recombination as well as F2 recombination, was used to narrow a QTL for alcohol drinking to a 2 cM region on distal chromosome 10 that was in common in the HAD1/LAD1 and HAD2/LAD2 analyses. Quantitative real time PCR (qRT-PCR) was used to examine mRNA expression of 6 candidate genes (Crebbp, Trap1, Gnptg, Clcn7, Fahd1, and Mapk8ip3) located within the narrowed QTL region in the HAD1/LAD1 rats. -

MAPK Kinase 3 Is a Tumor Suppressor with Reduced Copy Number in Breast Cancer
Author Manuscript Published OnlineFirst on November 14, 2013; DOI: 10.1158/0008-5472.CAN-13-1310 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. MAPK kinase 3 is a tumor suppressor with reduced copy number in breast cancer Adam J. MacNeil1,2,3, Shun-Chang Jiao4, Lori A. McEachern5, Yong Jun Yang6, Amanda Dennis1,2, Haiming Yu4, Zhaolin Xu3,7, Jean S. Marshall1,3, and Tong-Jun Lin1,2,3,7 1 Department of Microbiology & Immunology, Dalhousie University, Halifax, Nova Scotia, B3K 6R8, Canada. 2 Department of Pediatrics, Dalhousie University, Halifax, Nova Scotia, B3K 6R8, Canada. 3 Beatrice Hunter Cancer Research Institute, Suite 2L-A2, Tupper Link, 5850 College Street, PO Box 15000, Halifax, Nova Scotia, B3H 4R2, Canada. 4 Department of Medical Oncology, General Hospital of the People's Liberation Army, Beijing, 100853, China. 5 Department of Physiology & Biophysics, Dalhousie University, Halifax, Nova Scotia, B3K 6R8, Canada. 6 Institute of Zoonosis, College of Animal Sciences and Veterinary Medicine, Jilin University, Changchun, Jilin, 130062, China. 7 Department of Pathology, Dalhousie University, Halifax, Nova Scotia, B3K 6R8, Canada. Correspondence: Shun-Chang Jiao, email:[email protected]; Department of Medical Oncology, General Hospital of the People's Liberation Army, Beijing, 100853, China. Tong-Jun Lin, email: [email protected]; phone 902-470-8834; IWK Health Centre, Department of Pediatrics, Dalhousie University, 5850/5980 University Ave, Halifax, Nova Scotia, B3K 6R8, Canada. The authors have declared that no conflict of interest exists. Downloaded from cancerres.aacrjournals.org on September 29, 2021. © 2013 American Association for Cancer Research. -

(New Ref)-CG-MS
Send Orders for Reprints to [email protected] 522 Current Genomics, 2018, 19, 522-602 REVIEW ARTICLE Early Life Stress and Epigenetics in Late-onset Alzheimer’s Dementia: A Systematic Review Erwin Lemche* Section of Cognitive Neuropsychiatry, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK Abstract: Involvement of life stress in Late-Onset Alzheimer’s Disease (LOAD) has been evinced in longitudinal cohort epidemiological studies, and endocrinologic evidence suggests involvements of catecholamine and corticosteroid systems in LOAD. Early Life Stress (ELS) rodent models have suc- cessfully demonstrated sequelae of maternal separation resulting in LOAD-analogous pathology, thereby supporting a role of insulin receptor signalling pertaining to GSK-3beta facilitated tau hyper- phosphorylation and amyloidogenic processing. Discussed are relevant ELS studies, and findings from three mitogen-activated protein kinase pathways (JNK/SAPK pathway, ERK pathway, p38/MAPK pathway) relevant for mediating environmental stresses. Further considered were the roles of auto- phagy impairment, neuroinflammation, and brain insulin resistance. For the meta-analytic evaluation, 224 candidate gene loci were extracted from reviews of animal stud- ies of LOAD pathophysiological mechanisms, of which 60 had no positive results in human LOAD association studies. These loci were combined with 89 gene loci confirmed as LOAD risk genes in A R T I C L E H I S T O R Y previous GWAS and WES. Of the 313 risk gene loci evaluated, there were 35 human reports on epi- Received: July 01, 2017 genomic modifications in terms of methylation or histone acetylation. 64 microRNA gene regulation Revised: July 27, 2017 mechanisms were published for the compiled loci. -

Integrated Molecular Characterisation of the MAPK Pathways in Human
ARTICLE https://doi.org/10.1038/s42003-020-01552-6 OPEN Integrated molecular characterisation of the MAPK pathways in human cancers reveals pharmacologically vulnerable mutations and gene dependencies 1234567890():,; ✉ Musalula Sinkala 1 , Panji Nkhoma 2, Nicola Mulder 1 & Darren Patrick Martin1 The mitogen-activated protein kinase (MAPK) pathways are crucial regulators of the cellular processes that fuel the malignant transformation of normal cells. The molecular aberrations which lead to cancer involve mutations in, and transcription variations of, various MAPK pathway genes. Here, we examine the genome sequences of 40,848 patient-derived tumours representing 101 distinct human cancers to identify cancer-associated mutations in MAPK signalling pathway genes. We show that patients with tumours that have mutations within genes of the ERK-1/2 pathway, the p38 pathways, or multiple MAPK pathway modules, tend to have worse disease outcomes than patients with tumours that have no mutations within the MAPK pathways genes. Furthermore, by integrating information extracted from various large-scale molecular datasets, we expose the relationship between the fitness of cancer cells after CRISPR mediated gene knockout of MAPK pathway genes, and their dose-responses to MAPK pathway inhibitors. Besides providing new insights into MAPK pathways, we unearth vulnerabilities in specific pathway genes that are reflected in the re sponses of cancer cells to MAPK targeting drugs: a revelation with great potential for guiding the development of innovative therapies. -

Temperature Regulates NF-Κb Dynamics and Function Through Timing of A20 Transcription
Temperature regulates NF-κB dynamics and function through timing of A20 transcription C. V. Harpera,1, D. J. Woodcockb,c,1, C. Lama, M. Garcia-Albornozd, A. Adamsona, L. Ashalla, W. Rowea, P. Downtona, L. Schmidta, S. Weste, D. G. Spillera, D. A. Randb,c,2, and M. R. H. Whitea,2 aSystems Microscopy Centre, Division of Molecular and Cellular Function, School of Biology, Faculty of Biology, Medicine and Health, Manchester Academic Health Sciences Centre, University of Manchester, M13 9PT Manchester, United Kingdom; bMathematics Institute, University of Warwick, CV4 7AL Coventry, United Kingdom; cZeeman Institute for Systems Biology and Infectious Epidemiology Research, University of Warwick, CV4 7AL Coventry, United Kingdom; dDivision of Evolution and Genomic Sciences, School of Biology, Faculty of Biology, Medicine and Health, Manchester Academic Health Sciences Centre, University of Manchester, M13 9PT Manchester, United Kingdom; and eInstitute of Integrative Biology, University of Liverpool, L69 7ZB United Kingdom Edited by Ronald N. Germain, National Institutes of Health, Bethesda, MD, and approved April 27, 2018 (received for review March 7, 2018) NF-κB signaling plays a pivotal role in control of the inflammatory by measuring both the oscillatory dynamics and the downstream gene response. We investigated how the dynamics and function of NF-κB expression response. were affected by temperature within the mammalian physiological range (34 °C to 40 °C). An increase in temperature led to an increase Results in NF-κB nuclear/cytoplasmic oscillation frequency following Tumor To investigate the effect of temperature on NF-κB dynamics, we Necrosis Factor alpha (TNFα) stimulation. Mathematical modeling used time-lapse live-cell microscopy to study cytoplasmic to nu- suggested that this temperature sensitivity might be due to an clear oscillations of NF-κB across the physiological temper- A20-dependent mechanism, and A20 silencing removed the sensi- ature range, 34 °C to 40 °C. -
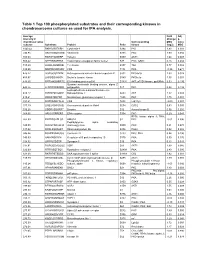
Table 1 Top 100 Phosphorylated Substrates and Their Corresponding Kinases in Chondrosarcoma Cultures As Used for IPA Analysis
Table 1 Top 100 phosphorylated substrates and their corresponding kinases in chondrosarcoma cultures as used for IPA analysis. Average Fold Adj intensity in Change p- chondrosarcoma Corresponding MSC value cultures Substrate Protein Psite kinase (log2) MSC 1043.42 RKKKVSSTKRH Cytohesin-1 S394 PKC 1.83 0.001 746.95 RKGYRSQRGHS Vitronectin S381 PKC 1.00 0.056 709.03 RARSTSLNERP Tuberin S939 AKT1 1.64 0.008 559.42 SPPRSSLRRSS Transcription elongation factor A-like1 S37 PKC; GSK3 0.18 0.684 515.29 LRRSLSRSMSQ Telethonin S157 Titin 0.77 0.082 510.00 MQPDNSSDSDY CD5 T434 PKA -0.35 0.671 476.27 GGRGGSRARNL Heterogeneous nuclear ribonucleoprotein K S302 PKCdelta 1.03 0.028 455.97 LKPGSSHRKTK Bruton's tyrosine kinase S180 PKCbeta 1.55 0.001 444.65 RRRMASMQRTG E1A binding protein p300 S1834 AKT; p70S6 kinase; pp90Rsk 0.53 0.195 Guanine nucleotide binding protein, alpha Z 440.26 HLRSESQRQRR polypeptide S27 PKC 0.88 0.199 6-phosphofructo-2-kinase/fructose-2,6- 424.12 RPRNYSVGSRP biphosphatase 2 S483 AKT 1.32 0.003 419.61 KKKIATRKPRF Metabotropic glutamate receptor 1 T695 PKC 1.75 0.001 391.21 DNSSDSDYDLH CD5 T453 Lck; Fyn -2.09 0.001 377.39 LRQLRSPRRAQ Ras associated protein Rab4 S204 CDC2 0.63 0.091 376.28 SSQRVSSYRRT Desmin S12 Aurora kinase B 0.56 0.255 369.05 ARIGGSRRERS EP4 receptor S354 PKC 0.29 0.543 RPS6 kinase alpha 3; PKA; 367.99 EPKRRSARLSA HMG14 S7 PKC -0.01 0.996 Peptidylglycine alpha amidating 349.08 SRKGYSRKGFD monooxygenase S930 PKC 0.21 0.678 347.92 RRRLSSLRAST Ribosomal protein S6 S236 PAK2 0.02 0.985 346.84 RSNPPSRKGSG Connexin -

Galectin-1 Promotes Lung Cancer Progression and Chemoresistance by Upregulating P38 MAPK, ERK, and Cyclooxygenase-2
Published OnlineFirst June 13, 2012; DOI: 10.1158/1078-0432.CCR-11-3348 Clinical Cancer Human Cancer Biology Research Galectin-1 Promotes Lung Cancer Progression and Chemoresistance by Upregulating p38 MAPK, ERK, and Cyclooxygenase-2 Ling-Yen Chung1, Shye-Jye Tang6, Guang-Huan Sun3, Teh-Ying Chou4, Tien-Shun Yeh2, Sung-Liang Yu5, and Kuang-Hui Sun1 Abstract Purpose: This study is aimed at investigating the role and novel molecular mechanisms of galectin-1 in lung cancer progression. Experimental Design: The role of galectin-1 in lung cancer progression was evaluated both in vitro and in vivo by short hairpin RNA (shRNA)-mediated knockdown of galectin-1 in lung adenocarcinoma cell lines. To explore novel molecular mechanisms underlying galectin-1–mediated tumor progression, we analyzed gene expression profiles and signaling pathways using reverse transcription PCR and Western blotting. A tissue microarray containing samples from patients with lung cancer was used to examine the expression of galectin-1 in lung cancer. Results: We found overexpression of galectin-1 in non–small cell lung cancer (NSCLC) cell lines. Suppression of endogenous galectin-1 in lung adenocarcinoma resulted in reduction of the cell migration, invasion, and anchorage-independent growth in vitro and tumor growth in mice. In particular, COX-2 was downregulated in galectin-1–knockdown cells. The decreased tumor invasion and anchorage-independent growth abilities were rescued after reexpression of COX-2 in galectin-1–knockdown cells. Furthermore, we found that TGF-b1 promoted COX-2 expression through galectin-1 interaction with Ras and subsequent activation of p38 mitogen-activated protein kinase (MAPK), extracellular signal–regulated kinase (ERK), and NF-kB pathway. -

Roles of P38α and P38β Mitogen‑Activated Protein Kinase Isoforms in Human Malignant Melanoma A375 Cells
INTERNATIONAL JOURNAL OF MOleCular meDICine 44: 2123-2132, 2019 Roles of p38α and p38β mitogen‑activated protein kinase isoforms in human malignant melanoma A375 cells SU-YING WEN1,2, SHI-YANN CHENG3,4, SHANG-CHUAN NG5, RITU ANEJA6, CHIH-JUNG CHEN7, CHIH-YANG HUANG8-12* and WEI-WEN KUO5* 1Department of Dermatology, Taipei City Hospital, Renai Branch, Taipei 106; 2Department of Health Care Management, National Taipei University of Nursing and Health Sciences, Taipei 112; 3Department of Medical Education and Research and Department of Obstetrics and Gynecology, China Medical University Beigang Hospital, Yunlin 65152; 4Obstetrics and Gynecology, School of Medicine, China Medical University; 5Department of Biological Science and Technology, College of Biopharmaceutical and Food Sciences, China Medical University, Taichung 404, Taiwan, R.O.C.; 6Department of Biology, Georgia State University, Atlanta, GA 30303, USA; 7Division of Breast Surgery, Department of Surgery, China Medical University Hospital; 8Graduate Institute of Biomedical Sciences, China Medical University, Taichung 404; 9Cardiovascular and Mitochondrial Related Disease Research Center, Hualien Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation; 10Center of General Education, Buddhist Tzu Chi Medical Foundation, Tzu Chi University of Science and Technology, Hualien 970; 11Department of Medical Research, China Medical University Hospital, China Medical University, Taichung 404; 12Department of Biotechnology, Asia University, Taichung 413, Taiwan, R.O.C. Received March 29, 2019; Accepted September 12, 2019 DOI: 10.3892/ijmm.2019.4383 Abstract. Skin cancer is one of the most common cancers vimentin, while mesenchymal-epithelial transition markers, worldwide. Melanoma accounts for ~5% of skin cancers but such as E-cadherin, were upregulated. Of note, the results causes the large majority of skin cancer-related deaths.