Molecular Characterization of Pseudomonas Syringae Pvs. from Different Host Plants by Repetitive Sequence-Based PCR and Multiplex-PCR
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Tekst Važećeg Statuta Sa Ugrađenim
Na osnovu člana 191. stav 1. Ustava Republike Srbije (“Službeni glasnik RS”, br. 98/2006) i člana 32. stav 1. tačka 1) Zakona o lokalnoj samoupravi (“Službeni glasnik RS”, br. 129/2007), Skupština grada Subotice, na 3. sednici održanoj dana 25.09.2008. godine, donela je STATUT GRADA SUBOTICE I OSNOVNE ODREDBE Član 1. Grad Subotica (u daljem tekstu: Grad) je teritorijalna jedinica u kojoj građani ostvaruju pravo na lokalnu samoupravu, u skladu sa Ustavom, zakonom i sa Statutom Grada Subotice (u daljem tekstu: Statut). Član 2. Teritoriju Grada, u skladu sa zakonom, čine naseljena mesta, odnosno područja katastarskih opština koje ulaze u njen sastav, i to: Redni Naziv naseljenog mesta Naziv naseljenog mesta na Naziv katastarske opštine broj na srpskom i hrvatskom mađarskom jeziku jeziku 1. Bajmok Bajmok Bajmok 2. Mišićevo Mišićevo Bajmok 3. Bački Vinogradi Királyhalom Bački Vinogradi 4. Bikovo Békova Bikovo 5. Gornji Tavankut Felsőtavankút Tavankut 6. Donji Tavankut Alsótavankút Tavankut 7. Ljutovo Mérges Tavankut 8. Hajdukovo Hajdújárás Palić 9. Šupljak Ludas Palić 10. Đurđin Györgyén Đurđin 11. Kelebija Kelebia Stari grad (deo) 12. Mala Bosna Kisbosznia Donji grad (deo) 13. Novi Žednik Újzsednik Žednik 14. Stari Žednik Nagyfény Žednik 15. Palić Palics Palić 16. Subotica Szabadka Donji grad (deo) Novi grad (deo) Stari grad (deo) 17. Čantavir Csantavér Čantavir 18. Bačko Dušanovo Dusanovó Čantavir 19. Višnjevac Visnyevác Čantavir Član 3. Naziv Grada je: GRAD SUBOTICA - na srpskom jeziku GRAD SUBOTICA - na hrvatskom jeziku SZABADKA VÁROS- na mađarskom jeziku. 1 Član 4. Sedište Grada je u Subotici, Trg slobode 1. Član 5. Grad ima svojstvo pravnog lica. -

Non-Technical Summary
NON-TECHNICAL SUMMARY According to the current regulation of categorization railway (“Official Gazette of RS", no. 75/2006) railroad Subotica (Cargo) - Horgoš - the Hungarian border is a regional railroad with the tag 56, and its length is 27.90 km. This railway has a category A, the maximum permissible weight of 16 t/axle, and the maximum weight per meter of 5.0 t/m '. Length stopping distance is 700 m. The railroad is single track and non-electrified. According to the ToR, the preliminary project of reconstruction and modernization of the railway line, within the limits of the railway land, which enable technical level and quality of traffic that is consistent with the established requirements of modern transport systems: • increase the speed of trains, • increasing axle loads, • increasing the length of trains, • increase line capacity, • increase safety on the line, • reducing the travel time of trains and energy consumption, • reducing the adverse environmental impact. In the investigated corridor of railway Subotica (Cargo) - Horgoš - Hungarian border potential of surface waters are natural waterways and melioration canals: Kereš – Radanovački canal, Tapša, Vinskipodrum, Kireš, Aranjšor, Dobo, Kamaraš and other melioration canals. Groundwater is particularly present in the lower parts of the terrain that are genetically related to the marsh sediments. The materials are mainly characterized by intergranular porosity, and depending on the amount of clay in individual members is conditioned their varying degree of permeability of the filtration coefficients of k = 10-4 - 10-7 cm / sec. The occurrence of groundwater was registered during the execution of research work at a depth of 0.90 to a depth of 1.90 m, extremely 4.20 m from terrain surface where drilling is performed. -

Treaty Series Recueil Des Traite's
Treaty Series Treaties and internationalagreements registered or filed and recorded with the Secretariat of the United Nations VOLUME 577 Recueil des Traite's Traitis et accords internationaux enregistres ou classes et inscrits au r'pertoire au Secritariat de l'Organisation des Nations Unies United Nations* Nations Unies New York, 1968 Treaties and internationalagreements registered or filed and recorded with the Secretariat of the United Nations VOLUME 577 1966 I. Nos. 8370-8381 TABLE OF CONTENTS Treaties and internationalagreements registeredfrom 9 November 1966 to 10 November 1966 Page No. 8370. Hungary and Yugoslavia: Agreement establishing regulations for the transport of goods by lorry or similar motor vehicle and the customs procedure in connexion there- with (with Protocol). Signed at Budapest, on 9 February 1962 .. ..... 3 No. 8371. Hungary and Yemen: Treaty of Friendship and Co-operation. Signed at Budapest, on 30 May 1964 . .. 39 No. 8372. Hungary and Yugoslavia: Convention concerning scientific, educational and cultural co-operation. Signed at Belgrade, on 15 October 1963 ... ............. .. 49 No. 8373. Hungary and Bulgaria: Agreement concerning scientific and cultural co-operation. Signed at Buda- pest, on 19 August 1965 ....... .................... ... 67 No. 8374. Hungary and Yugoslavia: Agreement concerning the abolition of the visa requirement. Signed at Budapest, on 23 November 1965 ..... ................ 89 No. 8375. Hungary and Yugoslavia: Agreement concerning the regulation of minor frontier traffic (with annexes). Signed at Budapest, on 9 August 1965 .... .............. ... 103 No. 8376. Hungary and Poland: Agreement concerning international motor transport. Signed at Budapest, on 18 July 1965 .... ... ... ......................... 161 Traitis et accords internationauxenregistris ou classis et inscrits au ripertoire au Secritariat de l'Organisationdes Nations Unies VOLUME 577 1966 I. -

Službeni List Br. 20
POŠTARINA PLAĆENA TISKOVINA KOD POŠTE 24000 SUBOTICA SLUŢBENI LIST GRADA SUBOTICE BROJ: 20 GODINA: LV DANA:26.jun 2019. CENA: 87,00 DIN. Republika Srbija AP Vojvodina Grad Subotica Skupština grada Subotice Izborna komisija Grada Subotice Broj: I-013-22/2019-1 Dana: 26.06.2019. godine Na osnovu ĉlana 28. Uputstva za sprovoĊenje izbora za ĉlanove saveta mesnih zajednica raspisanih za 7. jul 2019.god. („Sluţbeni list Grada Subotice“, br. 15/19), Izborna komisija Grada Subotice, na 27. sednici odrţanoj dana 26. juna 2019. godine, donela je ZBIRNU IZBORNU LISTU KANDIDATA ZA IZBOR ĈLANOVA SAVETA MESNE ZAJEDNICE ALEKSANDROVO I Zbirna izborna lista kandidata za izbor ĉlanova Saveta Mesne zajednice Aleksandrovo, raspisanih za 7. jul 2019. godine, sastoji se od sledećih izbornih lista sa sledećim kandidatima: 1.) Naziv izborne liste: SRPSKA NAPREDNA STRANKA - VAJDASÁGI MAGYAR SZÖVETSÉG Podnosilac izborne liste: SRPSKA NAPREDNA STRANKA - VAJDASÁGI MAGYAR SZÖVETSÉG Kandidati: 1. Bojan Nikolić, 1987, diplomirani ekonomista, Subotica, Aleksandrovi salaši 13 A 2. Vladimir Nješković (Vladimir Nješković), 1944, penzioner, Subotica, Deliblatska 13 3. Hajnalka Ileš (Hajnalka Ileš), 1973, nastavnik, Subotica, Plate Dobrojevića 7 4. Ljubica Rudić, 1952, penzioner, Subotica, Tolminska 1 A/2 5. Miroslav Vojnić Purĉar, 1988, higijeniĉar, Subotica, Gruţanska 4 6. Ankica Ĉukić, 1939, penzioner, Subotica, Plate Dobrojevića 17 7. Tibor Kuktin (Tibor Kuktin), 1959, elektrotehniĉar, Subotica, IĊoška 13 Strana 2 – Broj 20 Sluţbeni list Grada Subotice 26.jin 2019. 8. Vladimir Krstić (Vladimir Krstić), 1987, master informatike, Subotica, Vladimira Rolovića 22 9. Maja Skenderović, 1990, diplomirani ekonomista, Subotica, Aksentija Marodića 14 10. Tatjana Todorović, 1971, penzioner, Subotica, Mišić Borivoja 15 11. Angela Borbelj (Angela Borbelj), 1962, ekonomista, Subotica, Mišić Borivoja 69 12. -

Službeni List Grada Subotice
POŠTARINA PLA ĆENA TISKOVINA KOD POŠTE 24000 SUBOTICA SLUŽBENI LIST GRADA SUBOTICE BROJ: 24 GODINA: LI DANA:18. april 2016. CENA: 87,00 DIN. REPUBLIKA SRBIJA AP VOJVODINA GRAD SUBOTICA SKUPŠTINA GRADA SUBOTICE IZBORNA KOMISIJA GRADA SUBOTICE Broj: I-00-013-53/2016-4 Dana: 17. 4. 2016. godine Na osnovu člana 26. stav 1 i 2. Zakona o lokalnim izborima („Službeni glasnik RS“, broj 129/07, 34/10 – Odluka US i 54/11) Izborna komisija Grada Subotice, na 91.sednici održanoj dana 17.4.2016. godine u 22,35 časova utvrdila Pre čiš ćeni tekst Rešenja o utvr đivanju zbirne izborne liste za izbore za odbornike Skupštine grada Subotice raspisane za 24. april 2016. godine. Pre čiš ćeni tekst Rešenja o utvr đivanju zbirne izborne liste za izbore za odbornike Skupštine grada Subotice raspisane za 24. april 2016. godine obuhvata: 1. Rešenje o utvr đivanju zbirne izborne liste za izbore za odbornike Skupštine grada Subotice raspisane za 24. april 2016. godine („Službeni list Grada Subotice“, br. 21/16) izuzev ta čke II kojom je utvr đeno objavljivanje u „Službenom listu Grada Subotice“ i 2. Rešenje o dopuni Rešenja o utvr đivanju zbirne izborne liste za izbore za odbornike Skupštine grada Subotice raspisane za 24. april 2016. godine („Službeni list Grada Subotice“, br 23/16) izuzev ta čke II kojom je utvr đeno objavljivanje u „Službenom listu Grada Subotice“ i sa činjenje Pre čiš ćenog teksta Rešenja. REŠENJE O UTVR ĐIVANJU ZBIRNE IZBORNE LISTE ZA IZBORE ZA ODBORNIKE SKUPŠTINE GRADA SUBOTICE RASPISANE ZA 24. APRIL 2016. GODINE (pre čiš ćeni tekst) I Utvr đuje se Zbirna izborna lista, i to: 1. -

R.Br. Opština Mesto Adresa Prodavnice 1 Ada Obornjača Ada
R.br. Opština Mesto Najbliža prodavnica Adresa prodavnice 1 Ada Obornjača Ada Save Kovačevida 7 2 Aleksandrovac Bratidi Aleksandrovac 29. novembra 28 3 Aleksandrovac Bzenice Aleksandrovac 29. novembra 28 4 Aleksandrovac Koznica Aleksandrovac 29. novembra 28 5 Aleksandrovac Rogavčina Aleksandrovac 29. novembra 28 6 Aranđelovac Progoreoci Aranđelovac Knjaza Miloša 182 7 Babušnica Bogdanovac Vlasotince Nemanjina 47 8 Babušnica Ostatovica Vlasotince Nemanjina 47 9 Bačka Topola Bagremovo Bačka Topola Glavna 6 10 Bačka Topola Bogaraš Bačka Topola Glavna 6 11 Bačka Topola Kavilo Bačka Topola Glavna 6 12 Bačka Topola Midunovo Bačka Topola Glavna 6 13 Bačka Topola Obornjača Bačka Topola Glavna 6 14 Bačka Topola Srednji Salaš Bačka Topola Glavna 6 15 Bačka Topola Zobnatica Bačka Topola Glavna 6 16 Bajina Bašta Mala Reka Bajina bašta Vuka Karadzida 17 17 Bajina Bašta Zaugline Bajina bašta Vuka Karadzida 17 18 Bela Crkva Kaluđerovo Vršac Trg pobede 4 19 Bela Palanka Crveni Breg Pirot Trg pirotskih oslobodilaca bb 20 Bela Palanka Dolac (selo) Pirot Trg pirotskih oslobodilaca bb 21 Bela Palanka Donja Glama Pirot Trg pirotskih oslobodilaca bb 22 Bela Palanka Gornji Rinj Pirot Trg pirotskih oslobodilaca bb 23 Bela Palanka Leskovik Pirot Trg pirotskih oslobodilaca bb 24 Bela Palanka Telovac Pirot Trg pirotskih oslobodilaca bb 25 Bojnik Borince Lebane Cara Dušana, lamela 2 26 Bojnik Dobra Voda Lebane Cara Dušana, lamela 2 27 Bojnik Gornje Brijanje Lebane Cara Dušana, lamela 2 28 Bojnik Ivanje Lebane Cara Dušana, lamela 2 29 Bojnik Obražda Lebane Cara Dušana, lamela 2 30 Bojnik Orane Lebane Cara Dušana, lamela 2 31 Bosilegrad Brankovci Surdulica Kralja Petra 24 32 Bosilegrad Gložje Surdulica Kralja Petra 24 33 Bosilegrad Grujinci Surdulica Kralja Petra 24 34 Bosilegrad Izvor Surdulica Kralja Petra 24 35 Bosilegrad Milevci Surdulica Kralja Petra 24 36 Bosilegrad Zli Dol Surdulica Kralja Petra 24 37 Brus Grad Aleksandrovac 29. -

Master's Thesis
MASTER’S THESIS Planning Law and its Implementation in Serbia Planning Practice in Petrovac na Mlavi and Subotica Assistant Prof. Dipl.-Ing. Dr. Thomas Dillinger E280/7, Center of Regional Planning and Regional Development Department of Spatial Planning Vienna University of Technology Faculty of Architecture and Planning Robert Kolerovic, BSc Student ID: 0626019 Hetzendorferstraße 48 1120 Vienna Vienna, 3rd November 2014 Abstract Serbian cities and municipalities are facing many challenges when adopting spatial and urban plans and implementing them. One aspect, which is often mentioned by Serbian planners, is that the implementation of legal obligations, which are provided mainly by the 2009 Law on Planning and Construction, is insufficient. Another main aspect is that the degree of plan implementation is not very high. Both aspects lead to the hypothesis that the adoption of spatial and urban plans is done formally, especially because of the legal obligation, but the role and importance of the plans is neglected in reality, which is why plan implementation seems to be poor. The framework and preconditions of the case studies Subotica and Petrovac na Mlavi are different which is intended. Subotica is a city in the very north of the rather prosperous Autonomous Region of Vojvodina. Petrovac na Mlavi on the other hand is a municipality close to the Romanian border in the east of Serbia and is strongly influenced by agricultural structures. It can be concluded that smaller municipalities like Petrovac seem to have more problems with adopting and implementing plans, especially in regard to capacities of planning staff. The local level, i.e. -

Službeni List Opštine Subotica
POŠTARINA PLAĆENA TISKOVINA KOD POŠTE 24000 SUBOTICA SLUŽBENI LIST OPŠTINE SUBOTICA BROJ: 17 GODINA: XLIII DANA: 06. jun 2007. CENA: 65,00 DIN. Na osnovu člana 24. i člana 32. Odluke o mesnim zajednicama (prečišćeni tekst) («Službeni list Opštine Subotica», broj 12/2007) i člana 24. Uputstva za sprovođenje izbora za članove skupštine mesnih zajednica («Službeni list Opštine Subotica», broj 14/2007), Opštinska izborna komisija opštine Subotica, na sednici održanoj dana 06. juna 2007. godine, donela je ZBIRNU IZBORNU LISTU KANDIDATA ZA IZBOR ČLANOVA SKUPŠTINE MESNE ZAJEDNICE BAJMOK I Zbirna izborna lista kandidata za izbor članova Skupštine Mesne zajednice Bajmok koji se održava 17. juna 2007. godine sastoji se od sledećih izbornih lista sa sledećim kandidatima: 1.) Naziv izborne liste: SAVEZ VOJVOĐANSKIH MAĐARA – MAGLAI JENE Podnosilac izborne liste: Savez vojvođanskih Mađara Kandidati: 1. Maglai Jene, 1964., advokat, Bajmok, Beogradska 22 2. Hegediš Jožef, 1969. , veterinar, Bajmok, Vojvođanska 2 3. Šumegi Anastazija, 1983. , stomatološki tehn., Bajmok, Doža Đerđa 14/a 4. Kiš Arpad, 1955. , preduzetnik, Bajmok, Doža Đerđa 1 5. Penzeš Ištvan, 1949., mašinski inženjer, Bajmok, Trumbićeva 58/a 6. Ivanković Natalija, 1960., bankarski služb., Bajmok Ive Lole Ribara 30 7. Mojzeš Antal, 1934., penzioner, Bajmok, Moša Pijade 28 8. Klinocki Janoš, 1955., preduzetnik, Bajmok, Trumbićeva 27 9. Varga Matild, 1962., magacioner, Bajmok, Moša Pijade 29 10. Ritgaser Janoš, 1957, brusač, Bajmok, Frana Supila 34 11. Garajski Robert, 1974., poljoprivredni proizvođač, Bajmok, JNA 52 12. Pap Valerija, 1971., nastavnik razredne nastave, Bajmok, 9. Nova 10 13. Balaž Đerđ, 1967., poljoprivredni proizvođač, Bajmok, Čistac salaš 41 14. Poljaković Marija, 1960., penzioner, Bajmok, Subotički put 10 15. -

Srpski, Mađarski I Hrvatski
CIP – Каталогизација у публикацији Библиотека Матице српске , Нови Сад 502 . 175 ( 497 . 113 Subotica ) “2011“ МИТРОВИЋ, Сњежана Kvalitet životne sredine grada Subotice u 2011. godini / Snježana Mitrović, Ljiljana Krajnović, Iboja Farkaš ; [ prevodilac Čila Nemet ]. – Subotica : Otvoreni univerzitet, Regionalni Arhus centar, 2012 (Subotica : Printex). – 27, 27 Nasl. Str. Prištampanog prevoda: Az életkőrnyezet minősége Szabadka városában 2011-ben – Izvorni tekst i prevod štampani u međusobno obrnutim smerovima. – Tiraž 800. ISBN 978-86-87613-42-3 1. Kpајновић, Љиљана 2. Фаркаш, Ибоја a) Животна средина – Квалитет – Суботица - 2011 COBISS.SR-ID 269494791 1 Publikacija „Kvalitet životne sredine Grada Subotice u 2011. godini” je izdata u okviru projekta Regionalni Arhus centar Subotica Otvorenog univerziteta uz podršku Fonda za zaštitu životne sredine Republike Srbije i Grada Subotice. Izdavač: Otvoreni univerzitet Subotica – Regionalni Arhus centar Subotica Grad Subotica Za izdavača: Blažo Perović Grad Subotica Urednik izdanja: Pavle Budinčević Grad Subotica Autori: Snježana Mitrović Ljiljana Krajnović Iboja Farkaš Obrađivači: - Gradska uprava - Služba za zaštitu životne sredine i održivi razvoj mr Gordana Gavrilović i dipl. biolog Žika Reh - Zavod za javno zdravlje Subotica dr Zorica Mamužić Kukić i mr Nataša Čamprag Sabo Prevodilac: Čila Nemet Fotografije: Biljana Vučković Bence Mikeš Grad Subotica Oto Sekereš Dizajn i tehnička priprema: Agencija Organizator Štampa: Printex, Subotica Tiraž: 800 ISBN: 978-86-87613-42-3 Grad Subotica 2 Sadržaj -
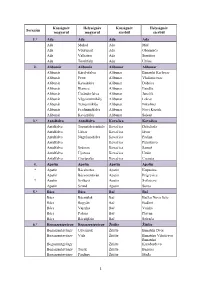
PDF Dokumentum
Községnév Helységnév Községnév Helységnév Sorszám magyarul magyarul szerbül szerbül 1.* Ada Ada Ada Ada Ada Mohol Ada Mol Ada Völgypart Ada Obornjača Ada Valkaisor Ada Sterijino Ada Törökfalu Ada Utrine 2. Alibunár Alibunár Alibunar Alibunar Alibunár Károlyfalva Alibunar Banatski Karlovac Alibunár Petre Alibunar Vladimirovac Alibunár Kevedobra Alibunar Dobrica Alibunár Illancsa Alibunar Ilandža Alibunár Újsándorfalva Alibunar Janošik Alibunár Végszentmihály Alibunar Lokve Alibunár Temesmiklós Alibunar Nikolinci Alibunár Ferdinándfalva Alibunar Novi Kozjak Alibunár Keviszőlős Alibunar Seleuš 3.* Antalfalva Antalfalva Kovačica Kovačica Antalfalva Torontálvásárhely Kovačica Debeljača Antalfalva Udvar Kovačica Idvor Antalfalva Nagylajosfalva Kovačica Padina Antalfalva Kovačica Putnikovo Antalfalva Számos Kovačica Samoš Antalfalva Újozora Kovačica Uzdin Antalfalva Cserépalja Kovačica Crepaja 4. Apatin Apatin Apatin Apatin * Apatin Bácskertes Apatin Kupusina Apatin Bácsszentiván Apatin Prigrevica * Apatin Szilágyi Apatin Svilojevo Apatin Szond Apatin Sonta 5.* Bács Bács Bač Bač Bács Bácsújlak Bač Bačko Novo Selo Bács Bogyán Bač Bođani Bács Vajszka Bač Vajska Bács Palona Bač Plavna Bács Bácsújfalu Bač Selenča 6.* Begaszentgyörgy Begaszentgyörgy Žitište Žitište Begaszentgyörgy Udvarnok Žitište Banatski Dvor Begaszentgyörgy Vida Žitište Banatsko Višnjićevo Banatsko Begaszentgyörgy Žitište Karađorđevo Begaszentgyörgy Torák Žitište Begejci Begaszentgyörgy Párdány Žitište Međa 1 Községnév Helységnév Községnév Helységnév Sorszám magyarul magyarul -

Korona Virus Novi Svinjski Grip
NOVE 13 ÉVE A POLGÁROK EGÉSZSÉGÉNEK SZOLGÁlatÁBAN Nikole Kujundžića br. 6 Radimo i subotom! DOLGOZUNK SZOMBATON IS! Godina II • Broj 151 • 31. januar 2020. • Cena: 40 dinara Subotičke novine istražuju da li se ponavlja 2009. godina kada smo milijarde dinara bacili na nepotrebne vakcine protiv nepostojeće bolesti Korona virus novi svinjski grip Kako je obična prehlada proglašena za smrtonosnu opasnost za planetu, opasniju od kuge str 7 Dobio posao zavarivača preko čitaoca Novih subotičkih novina!str 8 Zamena stare i dotrajalih uličnih sijalica u prigradskim naseljima donosi benefit 365004 Subotica na ISSN 2560-3655 uličnoj rasveti ISSN 2560-3655 772560 str 2 9 štedi milione evra 2 Broj 151 NOVE aktuelno 31. januar 2020. Zamena stare i dotrajalih uličnih sijalica u prigradskim naseljima donosi benefit Subotica na uličnoj rasveti štedi milione evra Novac koji bi grad morao da uloži u zamenu ra- svete sada može investirati u izgradnju novih i renoviranje postojećih vrtića, škola, domo- va zdravlja i ulagati u unapređenje komunal- ne infrastrukture godišnjem nivou iznositi 34 miliona dina- Postavljanje 12.914 novih LED sijalica Ljutovo, Mala Bosna, Mišićevo, Novi Žed- ra odnosno 70 odsto sredstava. Prema do- i prateće opreme u prigradskim naseljima nik, Stari Žednik, Nosa, Hajdukovo, Palić, sadašnjim rezultatima godišnja potrošnja uštedeće blizu četiri miliona evra u grad- Čantavir, Šupljak, Bačko Dušanovo, Bač- električne energije u postojećoj instalaci- skom budžetu. Godišnja ušteda kreće se ki Vinogradi, Mala Pešta, Đurđin, Kelebi- ji (sa trenutnom cenom električne energi- oko 350.000 evra, koje je grad nepotrebno ja i Gabrić. je koja sa naknadama iznosi 8.5 rsd) izno- trošio na plaćanje zastarele rasvete. -

Statut Grada Subotica
STATUT GRADA SUBOTICE ("Sl. list opštine Subotica", br. 26/2008 i 27/2008 - ispr.) I OSNOVNE ODREDBE Član 1 Grad Subotica (u daljem tekstu: Grad) je teritorijalna jedinica u kojoj gra đani ostvaruju pravo na lokalnu samoupravu, u skladu sa Ustavom, zakonom i sa Statutom Grada Subotice (u daljem tekstu: Statut). Član 2 Teritoriju Grada, u skladu sa zakonom, čine naseljena mesta, odnosno podru čja katastarskih opština koje ulaze u njen sastav, i to: Naziv naseljenog mesta Redni Naziv naseljenog mesta na srpskom i hrvatskom Naziv katastarske opštine broj na ma đarskom jeziku jeziku 1. Bajmok Bajmok Bajmok 2. Miši ćevo Miši ćevo Bajmok 3. Ba čki Vinogradi Királyhalom Ba čki Vinogradi 4. Bikovo Békova Bikovo 5. Gornji Tavankut Fels őtavankút Tavankut 6. Donji Tavankut Alsótavankút Tavankut 7. Ljutovo Mérges Tavankut 8. Hajdukovo Hajdújárás Pali ć 9. Šupljak Ludas Pali ć 10. Đur đin Györgyén Đur đin 11. Kelebija Kelebia Stari grad (deo) 12. Mala Bosna Kisbosznia Donji grad (deo) 13. Novi Žednik Újzsednik Žednik 14. Stari Žednik Nagyfény Žednik 15. Pali ć Palics Pali ć 16. Subotica Szabadka Donji grad (deo) Novi grad (deo) Stari grad (deo) 17. Čantavir Csantavér Čantavir 18. Ba čko Dušanovo Dusanovó Čantavir 19. Višnjevac Visnyevác Čantavir Član 3 Naziv Grada je: GRAD SUBOTICA - na srpskom jeziku GRAD SUBOTICA - na hrvatskom jeziku SZABADKA VÁROS - na ma đarskom jeziku. Član 4 Sedište Grada je u Subotici, Trg slobode 1. Član 5 Grad ima svojstvo pravnog lica. Član 6 Organi Grada imaju pe čat okruglog oblika sa tekstom: "Republika Srbija - Autonomna Pokrajina Vojvodina - Grad Subotica, naziv organa, Subotica" U sredini pe čata nalazi se grb Republike Srbije.