Summary of Excreted and Waterborne Viruses
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evidence for Viral Infection in the Copepods Labidocera Aestiva And
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School January 2012 Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida Darren Stephenson Dunlap University of South Florida, [email protected] Follow this and additional works at: http://scholarcommons.usf.edu/etd Part of the American Studies Commons, Other Oceanography and Atmospheric Sciences and Meteorology Commons, and the Virology Commons Scholar Commons Citation Dunlap, Darren Stephenson, "Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida" (2012). Graduate Theses and Dissertations. http://scholarcommons.usf.edu/etd/4032 This Thesis is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Evidence of Viruses in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida By Darren S. Dunlap A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science College of Marine Science University of South Florida Major Professor: Mya Breitbart, Ph.D Kendra Daly, Ph.D Ian Hewson, Ph.D Date of Approval: March 19, 2012 Key Words: Copepods, Single-stranded DNA Viruses, Mesozooplankton, Transmission Electron Microscopy, Metagenomics Copyright © 2012, Darren Stephenson Dunlap DEDICATION None of this would have been possible without the generous love and support of my entire family over the years. My parents, Steve and Jill Dunlap, have always encouraged my pursuits with support and love, and their persistence of throwing me into lakes and rivers is largely responsible for my passion for Marine Science. -

Seroprevalence of Antibodies to Primate Erythroparvovirus 1 (B19V) in Australia Helen M
Faddy et al. BMC Infectious Diseases (2018) 18:631 https://doi.org/10.1186/s12879-018-3525-7 RESEARCHARTICLE Open Access Seroprevalence of antibodies to primate erythroparvovirus 1 (B19V) in Australia Helen M. Faddy1,2* , Elise C. Gorman1,2, Veronica C. Hoad3, Francesca D. Frentiu2, Sarah Tozer4 and R. L. P. Flower1,2 Abstract Backgroud: Primate erythroparvovirus 1 (B19V) is a globally ubiquitous DNA virus. Infection results in a variety of clinical presentations including erythema infectiosum in children and arthralgia in adults. There is limited understanding of the seroprevalence of B19V antibodies in the Australian population and therefore of population- wide immunity. This study aimed to investigate the seroprevalence of B19V antibodies in an Australian blood donor cohort, along with a cohort from a paediatric population. Methods: Age/sex/geographical location stratified plasma samples (n = 2221) were collected from Australian blood donors. Samples were also sourced from paediatric patients (n = 223) in Queensland. All samples were screened for B19V IgG using an indirect- enzyme-linked immunosorbent assay. Results: Overall, 57.90% (95% CI: 55.94%–59.85%) of samples tested positive for B19V IgG, with the national age- standardized seroprevalence of B19V exposure in Australians aged 0 to 79 years estimated to be 54.41%. Increasing age (p < 0.001) and state of residence (p < 0.001) were independently associated with B19V exposure in blood donors, with the highest rates in donors from Tasmania (71.88%, 95% CI: 66.95%–76.80%) and donors aged 65–80 years (78.41%, 95% CI: 74.11%–82.71%). A seroprevalence of 52.04% (95% CI: 47.92%–56.15%) was reported in women of child-bearing age (16 to 44 years). -

Survival of Human Norovirus Surrogates in Juices and Their Inactivation Using Novel Methods
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Masters Theses Graduate School 5-2011 Survival of Human Norovirus Surrogates In Juices and their Inactivation Using Novel Methods Katie Marie Horm [email protected] Follow this and additional works at: https://trace.tennessee.edu/utk_gradthes Recommended Citation Horm, Katie Marie, "Survival of Human Norovirus Surrogates In Juices and their Inactivation Using Novel Methods. " Master's Thesis, University of Tennessee, 2011. https://trace.tennessee.edu/utk_gradthes/882 This Thesis is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Masters Theses by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a thesis written by Katie Marie Horm entitled "Survival of Human Norovirus Surrogates In Juices and their Inactivation Using Novel Methods." I have examined the final electronic copy of this thesis for form and content and recommend that it be accepted in partial fulfillment of the equirr ements for the degree of Master of Science, with a major in Food Science and Technology. Doris H. D'Souza, Major Professor We have read this thesis and recommend its acceptance: Federico M. Harte, Gina M. Pighetti Accepted for the Council: Carolyn R. Hodges Vice Provost and Dean of the Graduate School (Original signatures are on file with official studentecor r ds.) Survival of Human Norovirus Surrogates In Juices and their Inactivation Using Novel Methods A Thesis Presented for the Master of Science Degree The University of Tennessee, Knoxville Katie Marie Horm May 2011 Acknowledgments I would like to think my major professor/advisor Dr. -

Characterizing and Evaluating the Zoonotic Potential of Novel Viruses Discovered in Vampire Bats
viruses Article Characterizing and Evaluating the Zoonotic Potential of Novel Viruses Discovered in Vampire Bats Laura M. Bergner 1,2,* , Nardus Mollentze 1,2 , Richard J. Orton 2 , Carlos Tello 3,4, Alice Broos 2, Roman Biek 1 and Daniel G. Streicker 1,2 1 Institute of Biodiversity, Animal Health and Comparative Medicine, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8QQ, UK; [email protected] (N.M.); [email protected] (R.B.); [email protected] (D.G.S.) 2 MRC–University of Glasgow Centre for Virus Research, Glasgow G61 1QH, UK; [email protected] (R.J.O.); [email protected] (A.B.) 3 Association for the Conservation and Development of Natural Resources, Lima 15037, Peru; [email protected] 4 Yunkawasi, Lima 15049, Peru * Correspondence: [email protected] Abstract: The contemporary surge in metagenomic sequencing has transformed knowledge of viral diversity in wildlife. However, evaluating which newly discovered viruses pose sufficient risk of infecting humans to merit detailed laboratory characterization and surveillance remains largely speculative. Machine learning algorithms have been developed to address this imbalance by ranking the relative likelihood of human infection based on viral genome sequences, but are not yet routinely Citation: Bergner, L.M.; Mollentze, applied to viruses at the time of their discovery. Here, we characterized viral genomes detected N.; Orton, R.J.; Tello, C.; Broos, A.; through metagenomic sequencing of feces and saliva from common vampire bats (Desmodus rotundus) Biek, R.; Streicker, D.G. and used these data as a case study in evaluating zoonotic potential using molecular sequencing Characterizing and Evaluating the data. -

Enteric Viruses Nucleic Acids Distribution Along the Digestive Tract of Rhesus Macaques with Idiopathic Chronic Diarrhea
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.24.449827; this version posted June 24, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Enteric viruses nucleic acids distribution along the digestive tract of rhesus macaques with idiopathic chronic diarrhea Eric Delwart1,2*, David Merriam3,4, Amir Ardeshir3, Eda Altan1,2, Yanpeng Li1,2, Xutao Deng,1,2, J. Dennis Hartigan-O’Connor3 1. Vitlant Research Institute, 270 Masonic Ave, San Francisco CA94118 2. Dept of Laboratory Medicine, UCSF, San Francisco CA94118 3. California National Primate Research Center, University of California, Davis, CA 95616 4. Department of Pediatric Infectious Diseases, University of Colorado School of Medicine, Aurora, CO, USA. * Communicating author: [email protected] Abstract: Idiopathic chronic diarrhea (ICD) is a common clinical condition in captive rhesus macaques, claiming 33% of medical culls (i.e. deaths unrelated to research). Using viral metagenomics we characterized the eukaryotic virome in digestive tract tissues collected at necropsy from nine animals with ICD. We show the presence of multiple viruses in the Parvoviridae and Picornaviridae family. We then compared the distribution of viral reads in the stomach, duodenum, jejunum, ileum, and the proximal, transverse, and distal colons. Tissues and mucosal scraping from the same locations showed closely related results while different gut tissues from the same animal varied widely. Picornavirus reads were generally more abundant in the lower digestive tract, particularly in the descending (distal) colon. -

Opportunistic Intruders: How Viruses Orchestrate ER Functions to Infect Cells
REVIEWS Opportunistic intruders: how viruses orchestrate ER functions to infect cells Madhu Sudhan Ravindran*, Parikshit Bagchi*, Corey Nathaniel Cunningham and Billy Tsai Abstract | Viruses subvert the functions of their host cells to replicate and form new viral progeny. The endoplasmic reticulum (ER) has been identified as a central organelle that governs the intracellular interplay between viruses and hosts. In this Review, we analyse how viruses from vastly different families converge on this unique intracellular organelle during infection, co‑opting some of the endogenous functions of the ER to promote distinct steps of the viral life cycle from entry and replication to assembly and egress. The ER can act as the common denominator during infection for diverse virus families, thereby providing a shared principle that underlies the apparent complexity of relationships between viruses and host cells. As a plethora of information illuminating the molecular and cellular basis of virus–ER interactions has become available, these insights may lead to the development of crucial therapeutic agents. Morphogenesis Viruses have evolved sophisticated strategies to establish The ER is a membranous system consisting of the The process by which a virus infection. Some viruses bind to cellular receptors and outer nuclear envelope that is contiguous with an intri‑ particle changes its shape and initiate entry, whereas others hijack cellular factors that cate network of tubules and sheets1, which are shaped by structure. disassemble the virus particle to facilitate entry. After resident factors in the ER2–4. The morphology of the ER SEC61 translocation delivering the viral genetic material into the host cell and is highly dynamic and experiences constant structural channel the translation of the viral genes, the resulting proteins rearrangements, enabling the ER to carry out a myriad An endoplasmic reticulum either become part of a new virus particle (or particles) of functions5. -

ICTV Virus Taxonomy Profile: Parvoviridae
ICTV VIRUS TAXONOMY PROFILES Cotmore et al., Journal of General Virology 2019;100:367–368 DOI 10.1099/jgv.0.001212 ICTV ICTV Virus Taxonomy Profile: Parvoviridae Susan F. Cotmore,1,* Mavis Agbandje-McKenna,2 Marta Canuti,3 John A. Chiorini,4 Anna-Maria Eis-Hubinger,5 Joseph Hughes,6 Mario Mietzsch,2 Sejal Modha,6 Mylene Ogliastro,7 Judit J. Penzes, 2 David J. Pintel,8 Jianming Qiu,9 Maria Soderlund-Venermo,10 Peter Tattersall,1,11 Peter Tijssen12 and ICTV Report Consortium Abstract Members of the family Parvoviridae are small, resilient, non-enveloped viruses with linear, single-stranded DNA genomes of 4–6 kb. Viruses in two subfamilies, the Parvovirinae and Densovirinae, are distinguished primarily by their respective ability to infect vertebrates (including humans) versus invertebrates. Being genetically limited, most parvoviruses require actively dividing host cells and are host and/or tissue specific. Some cause diseases, which range from subclinical to lethal. A few require co-infection with helper viruses from other families. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the Parvoviridae, which is available at www.ictv.global/report/parvoviridae. Table 1. Characteristics of the family Parvoviridae Typical member: human parvovirus B19-J35 G1 (AY386330), species Primate erythroparvovirus 1, genus Erythroparvovirus, subfamily Parvovirinae Virion Small, non-enveloped, T=1 icosahedra, 23–28 nm in diameter Genome Linear, single-stranded DNA of 4–6 kb with short terminal hairpins Replication Rolling hairpin replication, a linear adaptation of rolling circle replication. Dynamic hairpin telomeres prime complementary strand and duplex strand-displacement synthesis; high mutation and recombination rates Translation Capped mRNAs; co-linear ORFs accessed by alternative splicing, non-consensus initiation or leaky scanning Host range Parvovirinae: mammals, birds, reptiles. -
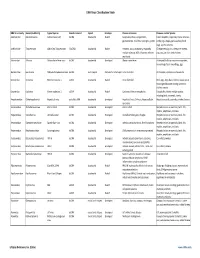
Virus Classification Tables V2.Vd.Xlsx
DNA Virus Classification Table DNA Virus Family Genera (Subfamily) Typical Species Genetic material Capsid Envelope Disease in Humans Diseases in other Species Adenoviridae Mastadenovirus Adenoviruses 1‐47 dsDNA Icosahedral Naked Respiratory illness; conjunctivitis, Canine hepatitis, respiratory illness in horses, gastroenteritis, tonsillitis, meningitis, cystitis cattle, pigs, sheep, goats, sea lions, birds dogs, squirrel enteritis Anelloviridae Torqueviruses Alpha‐Zeta Torqueviruses (‐)ssDNA Icosahedral Naked Hepatitis, lupus, pulmonary, myopathy, Chimpanzee, pig, cow, sheep, tree shrews, multiple sclerosis; 90% of humans infected pigs, cats, sea lions and chickens worldwide Asfarviridae Asfivirus African Swine fever virus dsDNA Icosahedral Enveloped African swine fever Arthropod (tick) transmission or ingestion; hemorrhagic fever in warthogs, pigs Baculoviridae Baculovirus Alpha‐Gamma Baculoviruses dsDNA Stick shaped Occluded or Enveloped none identified Arthropods, Lepidoptera, crustaceans Circoviridae Circovirus Porcine circovirus 1 ssDNA Icosahedral Naked none identified Birds, pigs, dogs; bats; rodents; causes post‐ weaning multisystem wasting syndrome, chicken anemia Circoviridae Cyclovirus Human cyclovirus 1 ssDNA Icosahedral Naked Cyclovirus Vietnam encephalitis Encephalitis; infects multiple species including birds, mammals, insects Hepadnaviridae Orthohepadnavirus Hepatitis B virus partially ssDNA Icosahedral Enveloped Hepatitis B virus; Cirrhosis, Hepatocellular Hepatitis in ducks, squirrels, primates, herons carcinoma Herpesviridae -

Viruses in Transplantation - Not Always Enemies
Viruses in transplantation - not always enemies Virome and transplantation ECCMID 2018 - Madrid Prof. Laurent Kaiser Head Division of Infectious Diseases Laboratory of Virology Geneva Center for Emerging Viral Diseases University Hospital of Geneva ESCMID eLibrary © by author Conflict of interest None ESCMID eLibrary © by author The human virome: definition? Repertoire of viruses found on the surface of/inside any body fluid/tissue • Eukaryotic DNA and RNA viruses • Prokaryotic DNA and RNA viruses (phages) 25 • The “main” viral community (up to 10 bacteriophages in humans) Haynes M. 2011, Metagenomic of the human body • Endogenous viral elements integrated into host chromosomes (8% of the human genome) • NGS is shaping the definition Rascovan N et al. Annu Rev Microbiol 2016;70:125-41 Popgeorgiev N et al. Intervirology 2013;56:395-412 Norman JM et al. Cell 2015;160:447-60 ESCMID eLibraryFoxman EF et al. Nat Rev Microbiol 2011;9:254-64 © by author Viruses routinely known to cause diseases (non exhaustive) Upper resp./oropharyngeal HSV 1 Influenza CNS Mumps virus Rhinovirus JC virus RSV Eye Herpes viruses Parainfluenza HSV Measles Coronavirus Adenovirus LCM virus Cytomegalovirus Flaviviruses Rabies HHV6 Poliovirus Heart Lower respiratory HTLV-1 Coxsackie B virus Rhinoviruses Parainfluenza virus HIV Coronaviruses Respiratory syncytial virus Parainfluenza virus Adenovirus Respiratory syncytial virus Coronaviruses Gastro-intestinal Influenza virus type A and B Human Bocavirus 1 Adenovirus Hepatitis virus type A, B, C, D, E Those that cause -

Diversity and Evolution of Viral Pathogen Community in Cave Nectar Bats (Eonycteris Spelaea)
viruses Article Diversity and Evolution of Viral Pathogen Community in Cave Nectar Bats (Eonycteris spelaea) Ian H Mendenhall 1,* , Dolyce Low Hong Wen 1,2, Jayanthi Jayakumar 1, Vithiagaran Gunalan 3, Linfa Wang 1 , Sebastian Mauer-Stroh 3,4 , Yvonne C.F. Su 1 and Gavin J.D. Smith 1,5,6 1 Programme in Emerging Infectious Diseases, Duke-NUS Medical School, Singapore 169857, Singapore; [email protected] (D.L.H.W.); [email protected] (J.J.); [email protected] (L.W.); [email protected] (Y.C.F.S.) [email protected] (G.J.D.S.) 2 NUS Graduate School for Integrative Sciences and Engineering, National University of Singapore, Singapore 119077, Singapore 3 Bioinformatics Institute, Agency for Science, Technology and Research, Singapore 138671, Singapore; [email protected] (V.G.); [email protected] (S.M.-S.) 4 Department of Biological Sciences, National University of Singapore, Singapore 117558, Singapore 5 SingHealth Duke-NUS Global Health Institute, SingHealth Duke-NUS Academic Medical Centre, Singapore 168753, Singapore 6 Duke Global Health Institute, Duke University, Durham, NC 27710, USA * Correspondence: [email protected] Received: 30 January 2019; Accepted: 7 March 2019; Published: 12 March 2019 Abstract: Bats are unique mammals, exhibit distinctive life history traits and have unique immunological approaches to suppression of viral diseases upon infection. High-throughput next-generation sequencing has been used in characterizing the virome of different bat species. The cave nectar bat, Eonycteris spelaea, has a broad geographical range across Southeast Asia, India and southern China, however, little is known about their involvement in virus transmission. -

Is the ZIKV Congenital Syndrome and Microcephaly Due to Syndemism with Latent Virus Coinfection?
viruses Review Is the ZIKV Congenital Syndrome and Microcephaly Due to Syndemism with Latent Virus Coinfection? Solène Grayo Institut Pasteur de Guinée, BP 4416 Conakry, Guinea; [email protected] or [email protected] Abstract: The emergence of the Zika virus (ZIKV) mirrors its evolutionary nature and, thus, its ability to grow in diversity or complexity (i.e., related to genome, host response, environment changes, tropism, and pathogenicity), leading to it recently joining the circle of closed congenital pathogens. The causal relation of ZIKV to microcephaly is still a much-debated issue. The identification of outbreak foci being in certain endemic urban areas characterized by a high-density population emphasizes that mixed infections might spearhead the recent appearance of a wide range of diseases that were initially attributed to ZIKV. Globally, such coinfections may have both positive and negative effects on viral replication, tropism, host response, and the viral genome. In other words, the possibility of coinfection may necessitate revisiting what is considered to be known regarding the pathogenesis and epidemiology of ZIKV diseases. ZIKV viral coinfections are already being reported with other arboviruses (e.g., chikungunya virus (CHIKV) and dengue virus (DENV)) as well as congenital pathogens (e.g., human immunodeficiency virus (HIV) and cytomegalovirus (HCMV)). However, descriptions of human latent viruses and their impacts on ZIKV disease outcomes in hosts are currently lacking. This review proposes to select some interesting human latent viruses (i.e., herpes simplex virus 2 (HSV-2), Epstein–Barr virus (EBV), human herpesvirus 6 (HHV-6), human parvovirus B19 (B19V), and human papillomavirus (HPV)), whose virological features and Citation: Grayo, S. -

Cellular Entry and Uncoating of Naked and Quasi-Enveloped Human
RESEARCH ARTICLE Cellular entry and uncoating of naked and quasi-enveloped human hepatoviruses Efraı´nE Rivera-Serrano1,2, Olga Gonza´ lez-Lo´ pez1,2, Anshuman Das2, Stanley M Lemon2,3* 1Lineberger Comprehensive Cancer Center, The University of North Carolina at Chapel Hill, Chapel Hill, United States; 2Department of Medicine, The University of North Carolina at Chapel Hill, Chapel Hill, United States; 3Department of Microbiology and Immunology, The University of North Carolina at Chapel Hill, Chapel Hill, United States Abstract Many ‘non-enveloped’ viruses, including hepatitis A virus (HAV), are released non- lytically from infected cells as infectious, quasi-enveloped virions cloaked in host membranes. Quasi-enveloped HAV (eHAV) mediates stealthy cell-to-cell spread within the liver, whereas stable naked virions shed in feces are optimized for environmental transmission. eHAV lacks virus- encoded surface proteins, and how it enters cells is unknown. We show both virion types enter by clathrin- and dynamin-dependent endocytosis, facilitated by integrin b1, and traffic through early and late endosomes. Uncoating of naked virions occurs in late endosomes, whereas eHAV undergoes ALIX-dependent trafficking to lysosomes where the quasi-envelope is enzymatically degraded and uncoating ensues coincident with breaching of endolysosomal membranes. Neither virion requires PLA2G16, a phospholipase essential for entry of other picornaviruses. Thus naked and quasi-enveloped virions enter via similar endocytic pathways, but uncoat in different compartments and release their genomes to the cytosol in a manner mechanistically distinct from other Picornaviridae. DOI: https://doi.org/10.7554/eLife.43983.001 *For correspondence: [email protected] Competing interests: The Introduction authors declare that no The presence or absence of an external lipid envelope has featured strongly in the systematic classi- competing interests exist.