Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Materials: Evaluation of Cytotoxicity and Α-Glucosidase Inhibitory Activity of Amide and Polyamino-Derivatives of Lupane Triterpenoids
Supplementary Materials: Evaluation of cytotoxicity and α-glucosidase inhibitory activity of amide and polyamino-derivatives of lupane triterpenoids Oxana B. Kazakova1*, Gul'nara V. Giniyatullina1, Akhat G. Mustafin1, Denis A. Babkov2, Elena V. Sokolova2, Alexander A. Spasov2* 1Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences, 71, pr. Oktyabrya, 450054 Ufa, Russian Federation 2Scientific Center for Innovative Drugs, Volgograd State Medical University, Novorossiyskaya st. 39, Volgograd 400087, Russian Federation Correspondence Prof. Dr. Oxana B. Kazakova Ufa Institute of Chemistry of the Ufa Federal Research Centre of the Russian Academy of Sciences 71 Prospeсt Oktyabrya Ufa, 450054 Russian Federation E-mail: [email protected] Prof. Dr. Alexander A. Spasov Scientific Center for Innovative Drugs of the Volgograd State Medical University 39 Novorossiyskaya st. Volgograd, 400087 Russian Federation E-mail: [email protected] Figure S1. 1H and 13C of compound 2. H NH N H O H O H 2 2 Figure S2. 1H and 13C of compound 4. NH2 O H O H CH3 O O H H3C O H 4 3 Figure S3. Anticancer screening data of compound 2 at single dose assay 4 Figure S4. Anticancer screening data of compound 7 at single dose assay 5 Figure S5. Anticancer screening data of compound 8 at single dose assay 6 Figure S6. Anticancer screening data of compound 9 at single dose assay 7 Figure S7. Anticancer screening data of compound 12 at single dose assay 8 Figure S8. Anticancer screening data of compound 13 at single dose assay 9 Figure S9. Anticancer screening data of compound 14 at single dose assay 10 Figure S10. -

Rab3a As a Modulator of Homeostatic Synaptic Plasticity
Wright State University CORE Scholar Browse all Theses and Dissertations Theses and Dissertations 2014 Rab3A as a Modulator of Homeostatic Synaptic Plasticity Andrew G. Koesters Wright State University Follow this and additional works at: https://corescholar.libraries.wright.edu/etd_all Part of the Biomedical Engineering and Bioengineering Commons Repository Citation Koesters, Andrew G., "Rab3A as a Modulator of Homeostatic Synaptic Plasticity" (2014). Browse all Theses and Dissertations. 1246. https://corescholar.libraries.wright.edu/etd_all/1246 This Dissertation is brought to you for free and open access by the Theses and Dissertations at CORE Scholar. It has been accepted for inclusion in Browse all Theses and Dissertations by an authorized administrator of CORE Scholar. For more information, please contact [email protected]. RAB3A AS A MODULATOR OF HOMEOSTATIC SYNAPTIC PLASTICITY A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy By ANDREW G. KOESTERS B.A., Miami University, 2004 2014 Wright State University WRIGHT STATE UNIVERSITY GRADUATE SCHOOL August 22, 2014 I HEREBY RECOMMEND THAT THE DISSERTATION PREPARED UNDER MY SUPERVISION BY Andrew G. Koesters ENTITLED Rab3A as a Modulator of Homeostatic Synaptic Plasticity BE ACCEPTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF Doctor of Philosophy. Kathrin Engisch, Ph.D. Dissertation Director Mill W. Miller, Ph.D. Director, Biomedical Sciences Ph.D. Program Committee on Final Examination Robert E. W. Fyffe, Ph.D. Vice President for Research Dean of the Graduate School Mark Rich, M.D./Ph.D. David Ladle, Ph.D. F. Javier Alvarez-Leefmans, M.D./Ph.D. Lynn Hartzler, Ph.D. -

Ran Activation Assay Kit
Product Manual Ran Activation Assay Kit Catalog Number STA-409 20 assays FOR RESEARCH USE ONLY Not for use in diagnostic procedures Introduction Small GTP-binding proteins (or GTPases) are a family of proteins that serve as molecular regulators in signaling transduction pathways. Ran, a 25 kDa protein of the Ras superfamily, regulates a variety of biological response pathways that include DNA synthesis, cell cycle progression, and translocation of RNA/proteins through the nuclear pore complex. Like other small GTPases, Ran regulates molecular events by cycling between an inactive GDP-bound form and an active GTP-bound form. In its active (GTP-bound) state, Ran binds specifically to RanBP1 to control downstream signaling cascades. Cell Biolabs’ Ran Activation Assay Kit utilizes RanBP1 Agarose beads to selectively isolate and pull- down the active form of Ran from purified samples or endogenous lysates. Subsequently, the precipitated GTP-Ran is detected by western blot analysis using an anti-Ran antibody. Cell Biolabs’ Ran Activation Assay Kit provides a simple and fast tool to monitor the activation of Ran. The kit includes easily identifiable RanBP1 Agarose beads (see Figure 1), pink in color, and a GTPase Immunoblot Positive Control for quick Ran identification. Each kit provides sufficient quantities to perform 20 assays. Figure 1: RanBP1 Agarose beads, in color, are easy to visualize, minimizing potential loss during washes and aspirations. 2 Assay Principle Related Products 1. STA-400: Pan-Ras Activation Assay Kit 2. STA-400-H: H-Ras Activation Assay Kit 3. STA-400-K: K-Ras Activation Assay Kit 4. STA-400-N: N-Ras Activation Assay Kit 5. -

Podocyte Specific Knockdown of Klf15 in Podocin-Cre Klf15flox/Flox Mice Was Confirmed
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1: Podocyte specific knockdown of Klf15 in Podocin-Cre Klf15flox/flox mice was confirmed. (A) Primary glomerular epithelial cells (PGECs) were isolated from 12-week old Podocin-Cre Klf15flox/flox and Podocin-Cre Klf15+/+ mice and cultured at 37°C for 1 week. Real-time PCR was performed for Nephrin, Podocin, Synaptopodin, and Wt1 mRNA expression (n=6, ***p<0.001, Mann-Whitney test). (B) Real- time PCR was performed for Klf15 mRNA expression (n=6, *p<0.05, Mann-Whitney test). (C) Protein was also extracted and western blot analysis for Klf15 was performed. The representative blot of three independent experiments is shown in the top panel. The bottom panel shows the quantification of Klf15 by densitometry (n=3, *p<0.05, Mann-Whitney test). (D) Immunofluorescence staining for Klf15 and Wt1 was performed in 12-week old Podocin-Cre Klf15flox/flox and Podocin-Cre Klf15+/+ mice. Representative images from four mice in each group are shown in the left panel (X 20). Arrows show colocalization of Klf15 and Wt1. Arrowheads show a lack of colocalization. Asterisk demonstrates nonspecific Wt1 staining. “R” represents autofluorescence from RBCs. In the right panel, a total of 30 glomeruli were selected in each mouse and quantification of Klf15 staining in the podocytes was determined by the ratio of Klf15+ and Wt1+ cells to Wt1+ cells (n=6 mice, **p<0.01, unpaired t test). Supplementary Figure 2: LPS treated Podocin-Cre Klf15flox/flox mice exhibit a lack of recovery in proteinaceous casts and tubular dilatation after DEX administration. -

Human Placenta Exosomes: Biogenesis, Isolation, Composition, and Prospects for Use in Diagnostics
International Journal of Molecular Sciences Review Human Placenta Exosomes: Biogenesis, Isolation, Composition, and Prospects for Use in Diagnostics Evgeniya E. Burkova 1,* , Sergey E. Sedykh 1,2 and Georgy A. Nevinsky 1,2 1 SB RAS Institute of Chemical Biology and Fundamental Medicine, 630090 Novosibirsk, Russia; [email protected] (S.E.S.); [email protected] (G.A.N.) 2 Department of Natural Sciences, Novosibirsk State University, 630090 Novosibirsk, Russia * Correspondence: [email protected]; Tel.: +7-(383)-363-51-27 Abstract: Exosomes are 40–100 nm nanovesicles participating in intercellular communication and transferring various bioactive proteins, mRNAs, miRNAs, and lipids. During pregnancy, the placenta releases exosomes into the maternal circulation. Placental exosomes are detected in the maternal blood even in the first trimester of pregnancy and their numbers increase significantly by the end of pregnancy. Exosomes are necessary for the normal functioning of the placenta and fetal development. Effects of exosomes on target cells depend not only on their concentration but also on their intrinsic components. The biochemical composition of the placental exosomes may cause various complications of pregnancy. Some studies relate the changes in the composition of nanovesicles to placental dysfunction. Isolation of placental exosomes from the blood of pregnant women and the study of protein, lipid, and nucleic composition can lead to the development of methods for early diagnosis of pregnancy pathologies. This review describes the biogenesis of exosomes, methods Citation: Burkova, E.E.; Sedykh, S.E.; of their isolation, analyzes their biochemical composition, and considers the prospects for using Nevinsky, G.A. Human Placenta exosomes to diagnose pregnancy pathologies. -

Defining Functional Interactions During Biogenesis of Epithelial Junctions
ARTICLE Received 11 Dec 2015 | Accepted 13 Oct 2016 | Published 6 Dec 2016 | Updated 5 Jan 2017 DOI: 10.1038/ncomms13542 OPEN Defining functional interactions during biogenesis of epithelial junctions J.C. Erasmus1,*, S. Bruche1,*,w, L. Pizarro1,2,*, N. Maimari1,3,*, T. Poggioli1,w, C. Tomlinson4,J.Lees5, I. Zalivina1,w, A. Wheeler1,w, A. Alberts6, A. Russo2 & V.M.M. Braga1 In spite of extensive recent progress, a comprehensive understanding of how actin cytoskeleton remodelling supports stable junctions remains to be established. Here we design a platform that integrates actin functions with optimized phenotypic clustering and identify new cytoskeletal proteins, their functional hierarchy and pathways that modulate E-cadherin adhesion. Depletion of EEF1A, an actin bundling protein, increases E-cadherin levels at junctions without a corresponding reinforcement of cell–cell contacts. This unexpected result reflects a more dynamic and mobile junctional actin in EEF1A-depleted cells. A partner for EEF1A in cadherin contact maintenance is the formin DIAPH2, which interacts with EEF1A. In contrast, depletion of either the endocytic regulator TRIP10 or the Rho GTPase activator VAV2 reduces E-cadherin levels at junctions. TRIP10 binds to and requires VAV2 function for its junctional localization. Overall, we present new conceptual insights on junction stabilization, which integrate known and novel pathways with impact for epithelial morphogenesis, homeostasis and diseases. 1 National Heart and Lung Institute, Faculty of Medicine, Imperial College London, London SW7 2AZ, UK. 2 Computing Department, Imperial College London, London SW7 2AZ, UK. 3 Bioengineering Department, Faculty of Engineering, Imperial College London, London SW7 2AZ, UK. 4 Department of Surgery & Cancer, Faculty of Medicine, Imperial College London, London SW7 2AZ, UK. -
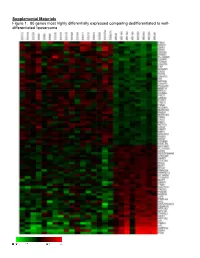
Supplemental Materials Figure 1. 80 Genes Most Highly Differentially Expressed Comparing Dedifferentiated to Well- Differentiated Liposarcoma
Supplemental Materials Figure 1. 80 genes most highly differentially expressed comparing dedifferentiated to well- differentiated liposarcoma Figure 2. Validation of microa CDC2, RACGAP1, FGFR-2, MAD2 and CITED1 200 CDK4 by Liposarcoma Subtype 16 0 12 0 80 Expression40 Level 0 Nl Fat rray data by quantitative RT-P Well Diff 16 0 p16 by Liposarcoma Subtype Dediff 12 0 80 Myxoid Expression40 Level Round Cell 0 12 0 MDM2 by Liposarcoma Subtype 10 0 Pleomorphic Nl Fat 80 60 40 Expression Level 20 RACGAP1 by LiposarcomaWell Diff Subtype 0 70 CR for genes CDK4, MDM2, p16, 60 Dediff 50 Nl Fat 40 30 Myxoid Expression20 Level Well Diff 10 50 0 Round Cell CDC2 by Liposarcoma Subtype Dediff 40 Nl Fat Pleomorphic 30 Myxoid 20 Expression Level Well Diff 10 MAD2L1 by Liposarcoma Subtype Round Cell 60 0 50 Dediff Pleomorphic 40 Nl Fat 30 Myxoid 20 Expression Level Well Diff 10 FGFR-2 by Liposarcoma Subtype 0 Round Cell 200 16 0 Dediff Nl Fat Pleomorphic 12 0 Myxoid 80 Expression Level Well Diff 40 Round Cell 0 Dediff Pleomorphic Nl Fat Myxoid Well Diff CITED1 by Liposarcoma Subtype Round Cell 10 0 80 Dediff Pleomorphic 60 40 Myxoid Expression Level 20 0 Round Cell Nl Fat Pleomorphic Well Diff Dediff Myxoid Round Cell Pleomorphic Table 1. 142 Gene classifier for liposarcoma subtype The genes used for each pair wise subtype comparison are grouped together. The flag column indicates which genes are unique to each subtype comparison. The values show the mean expression levels (actually the mean of the log expression levels was computed and than transformed back to absolute expression level). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Glioma Cell Secretion: a Driver of Tumor Progression and a Potential Therapeutic Target Damian A
Published OnlineFirst October 17, 2018; DOI: 10.1158/0008-5472.CAN-18-0345 Cancer Review Research Glioma Cell Secretion: A Driver of Tumor Progression and a Potential Therapeutic Target Damian A. Almiron Bonnin1,2, Matthew C. Havrda1,2, and Mark A. Israel1,2,3 Abstract Cellular secretion is an important mediator of cancer progres- ple oncogenic pathologies. In this review, we describe tumor cell sion. Secreted molecules in glioma are key components of secretion in high-grade glioma and highlight potential novel complex autocrine and paracrine pathways that mediate multi- therapeutic opportunities. Cancer Res; 78(21); 6031–9. Ó2018 AACR. Introduction Glioma-Secreted Molecules Impact Disease Glial cells in the central nervous system (CNS) provide trophic Progression support for neurons (1). In glial tumors, this trophic support is Glioma cells modify their microenvironment by introducing dysregulated creating a pro-oncogenic microenvironment medi- diverse molecules into the extracellular space (Table 1). To exem- ated by a heterogeneous array of molecules secreted into the plify the pro-oncogenic role that secreted molecules can have on – extracellular space (2 15). The glioma secretome includes pro- glioma pathology, we review the functional impact of specific teins, nucleic acids, and metabolites that are often overexpressed cytokines, metabolites, and nucleic acids on glioma biology. By in malignant tissue and contribute to virtually every aspect of describing some of the potent antitumorigenic effects observed in – cancer pathology (Table 1; Fig. 1; refs. 2 15), providing a strong preclinical therapeutic studies targeting tumor cell secretion, we – rationale to target the cancer cell secretory mechanisms. also highlight how blocking secreted molecules might be of fi Although the speci c mechanisms regulating secretion in therapeutic impact in gliomas, as well as other tumors. -

Supporting Information
Supporting Information Edgar et al. 10.1073/pnas.1601895113 SI Methods (Actimetrics), and recordings were analyzed using LumiCycle Mice. Sample size was determined using the resource equation: Data Analysis software (Actimetrics). E (degrees of freedom in ANOVA) = (total number of exper- – Cell Cycle Analysis of Confluent Cell Monolayers. NIH 3T3, primary imental animals) (number of experimental groups), with −/− sample size adhering to the condition 10 < E < 20. For com- WT, and Bmal1 fibroblasts were sequentially transduced − − parison of MuHV-4 and HSV-1 infection in WT vs. Bmal1 / with lentiviral fluorescent ubiquitin-based cell cycle indicators mice at ZT7 (Fig. 2), the investigator did not know the genotype (FUCCI) mCherry::Cdt1 and amCyan::Geminin reporters (32). of the animals when conducting infections, bioluminescence Dual reporter-positive cells were selected by FACS (Influx Cell imaging, and quantification. For bioluminescence imaging, Sorter; BD Biosciences) and seeded onto 35-mm dishes for mice were injected intraperitoneally with endotoxin-free lucif- subsequent analysis. To confirm that expression of mCherry:: Cdt1 and amCyan::Geminin correspond to G1 (2n DNA con- erin (Promega E6552) using 2 mg total per mouse. Following < ≤ anesthesia with isofluorane, they were scanned with an IVIS tent) and S/G2 (2 n 4 DNA content) cell cycle phases, Lumina (Caliper Life Sciences), 15 min after luciferin admin- respectively, cells were stained with DNA dye DRAQ5 (abcam) and analyzed by flow cytometry (LSR-Fortessa; BD Biosci- istration. Signal intensity was quantified using Living Image ences). To examine dynamics of replicative activity under ex- software (Caliper Life Sciences), obtaining maximum radiance perimental confluent conditions, synchronized FUCCI reporter for designated regions of interest (photons per second per − − − monolayers were observed by time-lapse live cell imaging over square centimeter per Steradian: photons·s 1·cm 2·sr 1), relative 3 d (Nikon Eclipse Ti-E inverted epifluorescent microscope). -

The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid
The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome Annette K. Brenner, Elise Aasebø, Maria Hernandez-Valladares, Frode Selheim, Frode Berven, Ida-Sofie Grønningsæter, Sushma Bartaula-Brevik and Øystein Bruserud Supplementary Material S2 of S31 Table S1. Detailed information about the 68 AML patients included in the study. # of blasts Viability Proliferation Cytokine Viable cells Change in ID Gender Age Etiology FAB Cytogenetics Mutations CD34 Colonies (109/L) (%) 48 h (cpm) secretion (106) 5 weeks phenotype 1 M 42 de novo 241 M2 normal Flt3 pos 31.0 3848 low 0.24 7 yes 2 M 82 MF 12.4 M2 t(9;22) wt pos 81.6 74,686 low 1.43 969 yes 3 F 49 CML/relapse 149 M2 complex n.d. pos 26.2 3472 low 0.08 n.d. no 4 M 33 de novo 62.0 M2 normal wt pos 67.5 6206 low 0.08 6.5 no 5 M 71 relapse 91.0 M4 normal NPM1 pos 63.5 21,331 low 0.17 n.d. yes 6 M 83 de novo 109 M1 n.d. wt pos 19.1 8764 low 1.65 693 no 7 F 77 MDS 26.4 M1 normal wt pos 89.4 53,799 high 3.43 2746 no 8 M 46 de novo 26.9 M1 normal NPM1 n.d. n.d. 3472 low 1.56 n.d. no 9 M 68 MF 50.8 M4 normal D835 pos 69.4 1640 low 0.08 n.d. -

Predicting Coupling Probabilities of G-Protein Coupled Receptors Gurdeep Singh1,2,†, Asuka Inoue3,*,†, J
Published online 30 May 2019 Nucleic Acids Research, 2019, Vol. 47, Web Server issue W395–W401 doi: 10.1093/nar/gkz392 PRECOG: PREdicting COupling probabilities of G-protein coupled receptors Gurdeep Singh1,2,†, Asuka Inoue3,*,†, J. Silvio Gutkind4, Robert B. Russell1,2,* and Francesco Raimondi1,2,* 1CellNetworks, Bioquant, Heidelberg University, Im Neuenheimer Feld 267, 69120 Heidelberg, Germany, 2Biochemie Zentrum Heidelberg (BZH), Heidelberg University, Im Neuenheimer Feld 328, 69120 Heidelberg, Germany, 3Graduate School of Pharmaceutical Sciences, Tohoku University, Sendai, Miyagi 980-8578, Japan and 4Department of Pharmacology and Moores Cancer Center, University of California, San Diego, La Jolla, CA 92093, USA Received February 10, 2019; Revised April 13, 2019; Editorial Decision April 24, 2019; Accepted May 01, 2019 ABSTRACT great use in tinkering with signalling pathways in living sys- tems (5). G-protein coupled receptors (GPCRs) control multi- Ligand binding to GPCRs induces conformational ple physiological states by transducing a multitude changes that lead to binding and activation of G-proteins of extracellular stimuli into the cell via coupling to situated on the inner cell membrane. Most of mammalian intra-cellular heterotrimeric G-proteins. Deciphering GPCRs couple with more than one G-protein giving each which G-proteins couple to each of the hundreds receptor a distinct coupling profile (6) and thus specific of GPCRs present in a typical eukaryotic organism downstream cellular responses. Determining these coupling is therefore critical to understand signalling. Here, profiles is critical to understand GPCR biology and phar- we present PRECOG (precog.russelllab.org): a web- macology. Despite decades of research and hundreds of ob- server for predicting GPCR coupling, which allows served interactions, coupling information is still missing for users to: (i) predict coupling probabilities for GPCRs many receptors and sequence determinants of coupling- specificity are still largely unknown.