Characterization of Shear-Sensitive Genes in the Normal Rat Aorta Identifies Hand2 As a Major Flow-Responsive Transcription Factor
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Uterine Double-Conditional Inactivation of Smad2 and Smad3 in Mice Causes Endometrial Dysregulation, Infertility, and Uterine Cancer
Uterine double-conditional inactivation of Smad2 and Smad3 in mice causes endometrial dysregulation, infertility, and uterine cancer Maya Krisemana,b, Diana Monsivaisa,c, Julio Agnoa, Ramya P. Masanda, Chad J. Creightond,e, and Martin M. Matzuka,c,f,g,h,1 aDepartment of Pathology and Immunology, Baylor College of Medicine, Houston, TX 77030; bReproductive Endocrinology and Infertility, Baylor College of Medicine/Texas Children’s Hospital Women’s Pavilion, Houston, TX 77030; cCenter for Drug Discovery, Baylor College of Medicine, Houston, TX 77030; dDepartment of Medicine, Baylor College of Medicine, Houston, TX 77030; eDan L. Duncan Comprehensive Cancer Center, Baylor College of Medicine, Houston, TX 77030; fDepartment of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030; gDepartment of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030; and hDepartment of Pharmacology and Chemical Biology, Baylor College of Medicine, Houston, TX 77030 Contributed by Martin M. Matzuk, December 6, 2018 (sent for review April 30, 2018; reviewed by Milan K. Bagchi and Thomas E. Spencer) SMAD2 and SMAD3 are downstream proteins in the transforming in endometrial function. Notably, members of the transforming growth factor-β (TGF β) signaling pathway that translocate signals growth factor β (TGF β) family are involved in many cellular from the cell membrane to the nucleus, bind DNA, and control the processes and serve as principal regulators of numerous biological expression of target genes. While SMAD2/3 have important roles functions, including female reproduction. Previous studies have in the ovary, we do not fully understand the roles of SMAD2/3 in shown the TGF β family to have key roles in ovarian folliculo- the uterus and their implications in the reproductive system. -

TGF-Β1 Signaling Targets Metastasis-Associated Protein 1, a New Effector in Epithelial Cells
Oncogene (2011) 30, 2230–2241 & 2011 Macmillan Publishers Limited All rights reserved 0950-9232/11 www.nature.com/onc ORIGINAL ARTICLE TGF-b1 signaling targets metastasis-associated protein 1, a new effector in epithelial cells SB Pakala1, K Singh1,3, SDN Reddy1, K Ohshiro1, D-Q Li1, L Mishra2 and R Kumar1 1Department of Biochemistry and Molecular Biology and Institute of Coregulator Biology, The George Washington University Medical Center, Washington, DC, USA and 2Department of Gastroenterology, Hepatology and Nutrition, The University of Texas MD Anderson Cancer Center, Houston, TX, USA In spite of a large number of transforming growth factor b1 gene chromatin in response to upstream signals. The (TGF-b1)-regulated genes, the nature of its targets with TGF-b1-signaling is largely mediated by Smad proteins roles in transformation continues to be poorly understood. (Massague et al., 2005) where Smad2 and Smad3 are Here, we discovered that TGF-b1 stimulates transcription phosphorylated by TGF-b1-receptors and associate with of metastasis-associated protein 1 (MTA1), a dual master the common mediator Smad4, which translocates to the coregulator, in epithelial cells, and that MTA1 status is a nucleus to participate in the expression of TGF-b1-target determinant of TGF-b1-induced epithelial-to-mesenchymal genes (Deckers et al., 2006). Previous studies have shown transition (EMT) phenotypes. In addition, we found that that CUTL1, also known as CDP (CCAAT displacement MTA1/polymerase II/activator protein-1 (AP-1) co-activator protein), a target of TGF-b1, is needed for its short-term complex interacts with the FosB-gene chromatin and stimu- effects of TGF-b1 on cell motility involving Smad4- lates its transcription, and FosB in turn, utilizes FosB/histone dependent pathway (Michl et al.,2005). -

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

Co-Occupancy by Multiple Cardiac Transcription Factors Identifies
Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart Aibin Hea,b,1, Sek Won Konga,b,c,1, Qing Maa,b, and William T. Pua,b,2 aDepartment of Cardiology and cChildren’s Hospital Informatics Program, Children’s Hospital Boston, Boston, MA 02115; and bHarvard Stem Cell Institute, Harvard University, Cambridge, MA 02138 Edited by Eric N. Olson, University of Texas Southwestern, Dallas, TX, and approved February 23, 2011 (received for review November 12, 2010) Identification of genomic regions that control tissue-specific gene study of a handful of model genes (e.g., refs. 7–10), it has not been expression is currently problematic. ChIP and high-throughput se- evaluated using unbiased, genome-wide approaches. quencing (ChIP-seq) of enhancer-associated proteins such as p300 In this study, we used a modified ChIP-seq approach to define identifies some but not all enhancers active in a tissue. Here we genome wide the binding sites of these cardiac TFs (1). We show that co-occupancy of a chromatin region by multiple tran- provide unbiased support for collaborative TF interactions in scription factors (TFs) identifies a distinct set of enhancers. GATA- driving cardiac gene expression and use this principle to show that chromatin co-occupancy by multiple TFs identifies enhancers binding protein 4 (GATA4), NK2 transcription factor-related, lo- with cardiac activity in vivo. The majority of these multiple TF- cus 5 (NKX2-5), T-box 5 (TBX5), serum response factor (SRF), and “ binding loci (MTL) enhancers were distinct from p300-bound myocyte-enhancer factor 2A (MEF2A), here referred to as cardiac enhancers in location and functional properties. -

Micrornas in Cardiovascular Disease: from Pathogenesis to Prevention and Treatment
MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment Daniel Quiat, Eric N. Olson J Clin Invest. 2013;123(1):11-18. https://doi.org/10.1172/JCI62876. Review Series The management of cardiovascular risk through lifestyle modification and pharmacotherapy is paramount to the prevention of cardiovascular disease. Epidemiological studies have identified obesity, dyslipidemia, diabetes, and hypertension as interrelated factors that negatively affect cardiovascular health. Recently, genetic and pharmacological evidence in model systems has implicated microRNAs as dynamic modifiers of disease pathogenesis. An expanded understanding of the function of microRNAs in gene regulatory networks associated with cardiovascular risk will enable identification of novel genetic mechanisms of disease and inform the development of innovative therapeutic strategies. Find the latest version: https://jci.me/62876/pdf Review series MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment Daniel Quiat and Eric N. Olson Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, Texas, USA. The management of cardiovascular risk through lifestyle modification and pharmacotherapy is paramount to the prevention of cardiovascular disease. Epidemiological studies have identified obesity, dyslipidemia, diabetes, and hypertension as interrelated factors that negatively affect cardiovascular health. Recently, genetic and pharmaco- logical evidence in model systems has implicated microRNAs -

HAND2‑Mediated Proteolysis Negatively Regulates the Function of Estrogen Receptor Α
5538 MOLECULAR MEDICINE REPORTS 12: 5538-5544, 2015 HAND2‑mediated proteolysis negatively regulates the function of estrogen receptor α TOMOHIKO FUKUDA*, AKIRA SHIRANE*, OSAMU WADA-HIRAIKE, KATSUTOSHI ODA, MICHIHIRO TANIKAWA, AYAKO SAKUABASHI, MANA HIRANO, HOUJU FU, YOSHIHIRO MORITA, YUICHIRO MIYAMOTO, KANAKO INABA, KEI KAWANA, YUTAKA OSUGA and TOMOYUKI FUJII Department of Obstetrics and Gynecology, Graduate School of Medicine, The University of Tokyo, Tokyo 113-8655, Japan Received October 2, 2014; Accepted June 11, 2015 DOI: 10.3892/mmr.2015.4070 Abstract. A previous study demonstrated that the transcriptional activation function of ERα was possibly attrib- progesterone-inducible HAND2 gene product is a basic uted to the proteasomic degradation of ERα by HAND2. These helix-loop-helix transcription factor and prevents mitogenic results indicate a novel anti-tumorigenic function of HAND2 effects of estrogen receptor α (ERα) by inhibiting fibroblast in regulating ERα-dependent gene expression. Considering growth factor signalling in mouse uteri. However, whether that HAND2 is commonly hypermethylated and silenced in HAND2 directly affects the transcriptional activation func- endometrial cancer, it is hypothesized that HAND2 may serve tion of ERα remains to be elucidated. In the present study, as a possible tumor suppressor, particularly in uterine tissue. the physical interaction between HAND2 and ERα was investigating by performing an immunoprecipitation assay Introduction and an in vitro pull-down assay. The results demonstrated that HAND2 and ERα interacted in a ligand-independent manner. Endometrial cancer is one of the most common types of The in vitro pull-down assays revealed a direct interaction gynecologic malignancy, increasing each year (1). Based on between HAND2 and the amino-terminus of ERα, termed a pathological view, endometrial cancer can be divided into the activation function-1 domain. -

PDF Hosted at the Radboud Repository of the Radboud University Nijmegen
PDF hosted at the Radboud Repository of the Radboud University Nijmegen The following full text is a publisher's version. For additional information about this publication click this link. http://hdl.handle.net/2066/147462 Please be advised that this information was generated on 2021-10-11 and may be subject to change. Chromatin Regulatory Complexes from an Interaction Proteomics Perspective an Interaction Proteomics from Regulatory Complexes Chromatin Chromatin Regulatory Complexes from an Interaction Proteomics Perspective H. İrem Baymaz H. İrem Baymaz H. İrem Baymaz 2015 Chromatin Regulatory Complexes from an Interaction Proteomics Perspective H. İrem Baymaz printed by: Proefschriftmaken.nl || Uitgeverij BOXPress Copyright © 2015 H. İrem Baymaz Chromatin Regulatory Complexes from an Interaction Proteomics Perspective Chromatine-Regulerende Complexen vanuit een Interactie-Proteomics Perspectief Proefschrift ter verkrijging van de graad van doctor aan de Radboud Universiteit Nijmegen op gezag van de rector magnificus prof. dr. Th.L.M. Engelen, volgens besluit van het college van decanen in het openbaar te verdedigen op donderdag 26 november 2015 om 12.30 uur precies door Hamdiye İrem Baymaz geboren op 8 maart 1986 te Ankara, Turkije Promoter: Prof. dr. M. Vermeulen Manuscriptcommissie: Prof. dr. J.H.L.M. van Bokhoven Prof. dr. H.T.M. Timmers (UMCU) Dr. L.M. Kamminga Anne ve Babama. Table of Contents List of Abbreviations 9 Chapter 1 11 Introduction Chapter 2 45 Identifying nuclear protein-protein interactions using GFP affinity purification -

I3C & Hormone Positive Breast Cancer
ARTICLE IN PRESS Journal of Nutritional Biochemistry xx (2006) xxx–xxx Estrogen receptor a as a target for indole-3-carbinol Thomas T.Y. Wanga,4, Matthew J. Milnerb, John A. Milnerc, Young S. Kimc aPhytonutrients Laboratory, Beltsville Human Nutrition Research Center, Agricultural Research Service, U.S. Department of Agriculture, Beltsville, MD 20705, USA bDepartment of Biology, The Pennsylvania State University, University Park, PA, USA cNutritional Science Research Group, Division of Cancer Prevention, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA Received 8 June 2005; received in revised form 14 October 2005; accepted 22 October 2005 Abstract A wealth of preclinical evidence supports the antitumorigenic properties of indole-3-carbinol (I3C), which is a major bioactive food component in cruciferous vegetables. However, the underlying molecular mechanism(s) accounting for these effects remain unresolved. In the present study, estrogen receptor alpha (ER-a) was identified as a potential molecular target for I3C. Treating MCF-7 cells with 100 AM I3C reduced ER-a mRNA expression by approximately 60% compared to controls. This reduction in ER-a transcript levels was confirmed using real-time polymerase chain reaction. The I3C dimer, 3,3V-diindolylmethane (DIM), was considerably more effective in depressing ER-a mRNA in MCF-7 cells than the monomeric unit. The suppressive effects of 5 AM DIM on ER-a mRNA was comparable to that caused by 100 AM I3C. DIM is known to accumulate in the nucleus and is a preferred ligand for aryl hydrocarbon receptor (AhR) to I3C. The addition of other AhR ligands, a-naphthoflavone (a-NF, 10 AM) and luteolin (10 AM), to the culture media resulted in a similar suppression in ER-a mRNA levels to that caused by 5 AM DIM. -
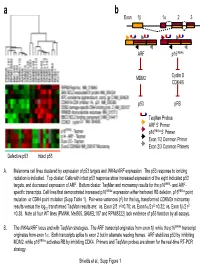
Taqman Probes ARF 5' Primer P16ink4a 5' Primer Exon 1/2
a b Exon 1β 1α 23 ARF p16INK4a MDM2 Cyclin D CDK4/6 p53 pRB TaqMan Probes ARF 5’ Primer p16INK4a 5’ Primer Exon 1/2 Common Primer Exon 2/3 Common Primers Defective p53 Intact p53 A. Melanoma cell lines clustered by expression of p53 targets and INK4a/ARF expression. The p53 response to ionizing radiation is indicated. Top cluster: Cells with intact p53 response show increased expression of the eight indicated p53 targets, and decreased expression of ARF. Bottom cluster: TaqMan and microarray results for the p16INK4- and ARF- specific transcripts. Cell lines that demonstrated increased p16INK4a expression either harbored RB deletion, p16INK4a point 2 mutation or CDK4 point mutation (Supp Table 1). Pair-wise variances (r ) for the log2 transformed CDKN2a microarray 2 2 2 results versus the log10 transformed TaqMan results are: vs. Exon 2/3 r =0.78; vs. Exon1α/2 r =0.32; vs. Exon 1β/2 r =0.38. Note all four WT lines (PMWK, Mel505, SKMEL187 and RPMI8322) lack evidence of p53 function by all assays. B. The INK4a/ARF locus and with TaqMan strategies. The ARF transcript originates from exon 1β while the p16INK4a transcript originates from exon 1α. Both transcripts splice to exon 2 but in alternate reading frames. ARF stabilizes p53 by inhibiting MDM2, while p16INK4a activates RB by inhibiting CDK4. Primers and TaqMan probes are shown for the real-time RT-PCR strategy. Shields et al., Supp Figure 1 SKMEL 28 U01 24h SKMEL WM2664 U01 48h WM2664 U01 24h 24 U01 48h SKMEL 24 U01 24h SKMEL 24 Untreated SKMEL 24 DMSO 48h SKMEL 24 DMSO 24h SKMEL WM2664 DMSO 24h WM2664 DMSO 48h WM2644 Untreated 28 Untreated SKMEL 28 DMSO 48h SKMEL 28 DMSO 24h SKMEL -3.00 -2.00 -1.00 0.00 1.00 2.00 3.00 relative to median expression Genes decreased by UO1 (863) HIF1A Hypoxia-inducible factor 1, alpha subunit NM_001530 RBBP8 Retinoblastoma binding protein 8 NM_002894 Homo sapiens, clone IMAGE:4337652, mRNA BC018676 EIF4EBP1 Eukaryotic translation initiation factor 4E binding protein EXOSC8 Exosome component 8 NM_181503 ENST00000321524 MCM7 MCM7 minichromosome maintenance deficient 7 (S. -

Genetic Factors in Nonsyndromic Orofacial Clefts
Published online: 2021-02-12 THIEME Review Article 101 Genetic Factors in Nonsyndromic Orofacial Clefts Mahamad Irfanulla Khan1 Prashanth C.S.2 Narasimha Murthy Srinath3 1 Department of Orthodontics & Dentofacial Orthopedics, The Oxford Address for correspondence Mahamad Irfanulla Khan, BDS, MDS, Dental College, Bangalore, Karnataka, India Department of Orthodontics & Dentofacial Orthopedics, The Oxford 2 Department of Orthodontics & Dentofacial Orthopedics, DAPM R.V. Dental College, Bangalore, Karnataka 560068, India Dental College, Bangalore, Karnataka, India (e-mail: [email protected]). 3 Department of Oral & Maxillofacial Surgery, Krishnadevaraya College of Dental Sciences, Bangalore, Karnataka, India Global Med Genet 2020;7:101–108. Abstract Orofacial clefts (OFCs) are the most common congenital birth defects in humans and immediately recognized at birth. The etiology remains complex and poorly understood and seems to result from multiple genetic and environmental factors along with gene– environment interactions. It can be classified into syndromic (30%) and nonsyndromic (70%) clefts. Nonsyndromic OFCs include clefts without any additional physical or Keywords cognitive deficits. Recently, various genetic approaches, such as genome-wide associ- ► orofacial clefts ation studies (GWAS), candidate gene association studies, and linkage analysis, have ► nonsyndromic identified multiple genes involved in the etiology of OFCs. ► genetics This article provides an insight into the multiple genes involved in the etiology of OFCs. ► gene mutation Identification of specific genetic causes of clefts helps in a better understanding of the ► genome-wide molecular pathogenesis of OFC. In the near future, it helps to provide a more accurate association study diagnosis, genetic counseling, personalized medicine for better clinical care, and ► linkage analysis prevention of OFCs. -

Nkx Genes Establish Second Heart Field Cardiomyocyte Progenitors At
© 2018. Published by The Company of Biologists Ltd | Development (2018) 145, dev161497. doi:10.1242/dev.161497 RESEARCH ARTICLE Nkx genes establish second heart field cardiomyocyte progenitors at the arterial pole and pattern the venous pole through Isl1 repression Sophie Colombo1, Carmen de Sena-Tomás1, Vanessa George1, Andreas A. Werdich2, Sunil Kapur2, Calum A. MacRae2 and Kimara L. Targoff1,* ABSTRACT Vincent and Buckingham, 2010; Zaffran et al., 2004). Errors in NKX2-5 is the most commonly mutated gene associated with human specification and differentiation of the SHF cells under these precise congenital heart defects (CHDs), with a predilection for cardiac pole spatial and temporal constraints can lead to a variety of congenital abnormalities. This homeodomain transcription factor is a central heart defects (CHDs), specifically conotruncal (OFT) and atrial regulator of cardiac development and is expressed in both the first (IFT) abnormalities. These malformations of the cardiac poles are and second heart fields (FHF and SHF). We have previously revealed found in 10% and 30%, respectively, of individuals with CHDs and essential functions of nkx2.5 and nkx2.7, two Nkx2-5 homologs result in significant neonatal morbidity and mortality (Gelb and expressed in zebrafish cardiomyocytes, in maintaining ventricular Chung, 2014; Hoffman and Kaplan, 2002; Hoffman et al., 2004; identity. However, the differential roles of these genes in the specific Loffredo, 2000; Nemer, 2008; Payne et al., 1995; Pierpont et al., subpopulations of the anterior (aSHF) and posterior (pSHF) SHFs 2007; Supino et al., 2006). Despite this clinical impact, the have yet to be fully defined. Here, we show that Nkx genes regulate mechanisms underlying the development of the arterial and venous aSHF and pSHF progenitors through independent mechanisms. -

Critical Roles of ARHGAP36 As a Signal Transduction Mediator of Shh
RESEARCH ARTICLE Critical roles of ARHGAP36 as a signal transduction mediator of Shh pathway in lateral motor columnar specification Heejin Nam1, Shin Jeon1,2, Hyejin An1, Jaeyoung Yoo1, Hyo-Jong Lee3, Soo-Kyung Lee2,4, Seunghee Lee1* 1College of Pharmacy and Research Institute of Pharmaceutical Sciences, Seoul National University, Seoul, Republic of Korea; 2Neuroscience Section, PapeFamily Pediatric Research Institute, Department of Pediatrics, Oregon Health and Science Uiversity, Portland, United States; 3College of Pharmacy and Inje Institute of Pharmaceutical Sciences and Research, Inje University, Gyungnam, Republic of Korea; 4Vollum Institute, Oregon Health and Science University, Portland, United States Abstract During spinal cord development, Sonic hedgehog (Shh), secreted from the floor plate, plays an important role in the production of motor neurons by patterning the ventral neural tube, which establishes MN progenitor identity. It remains unknown, however, if Shh signaling plays a role in generating columnar diversity of MNs that connect distinct target muscles. Here, we report that Shh, expressed in MNs, is essential for the formation of lateral motor column (LMC) neurons in vertebrate spinal cord. This novel activity of Shh is mediated by its downstream effector ARHGAP36, whose expression is directly induced by the MN-specific transcription factor complex Isl1-Lhx3. Furthermore, we found that AKT stimulates the Shh activity to induce LMC MNs through the stabilization of ARHGAP36 proteins. Taken together, our data reveal that Shh, secreted from MNs, plays a crucial role in generating MN diversity via a regulatory axis of Shh-AKT- ARHGAP36 in the developing mouse spinal cord. *For correspondence: DOI: https://doi.org/10.7554/eLife.46683.001 [email protected] Competing interests: The authors declare that no Introduction competing interests exist.