Genetic and Functional Differences Between Multipotent Neural and Pluripotent Embryonic Stem Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

TGF-Β1 Signaling Targets Metastasis-Associated Protein 1, a New Effector in Epithelial Cells
Oncogene (2011) 30, 2230–2241 & 2011 Macmillan Publishers Limited All rights reserved 0950-9232/11 www.nature.com/onc ORIGINAL ARTICLE TGF-b1 signaling targets metastasis-associated protein 1, a new effector in epithelial cells SB Pakala1, K Singh1,3, SDN Reddy1, K Ohshiro1, D-Q Li1, L Mishra2 and R Kumar1 1Department of Biochemistry and Molecular Biology and Institute of Coregulator Biology, The George Washington University Medical Center, Washington, DC, USA and 2Department of Gastroenterology, Hepatology and Nutrition, The University of Texas MD Anderson Cancer Center, Houston, TX, USA In spite of a large number of transforming growth factor b1 gene chromatin in response to upstream signals. The (TGF-b1)-regulated genes, the nature of its targets with TGF-b1-signaling is largely mediated by Smad proteins roles in transformation continues to be poorly understood. (Massague et al., 2005) where Smad2 and Smad3 are Here, we discovered that TGF-b1 stimulates transcription phosphorylated by TGF-b1-receptors and associate with of metastasis-associated protein 1 (MTA1), a dual master the common mediator Smad4, which translocates to the coregulator, in epithelial cells, and that MTA1 status is a nucleus to participate in the expression of TGF-b1-target determinant of TGF-b1-induced epithelial-to-mesenchymal genes (Deckers et al., 2006). Previous studies have shown transition (EMT) phenotypes. In addition, we found that that CUTL1, also known as CDP (CCAAT displacement MTA1/polymerase II/activator protein-1 (AP-1) co-activator protein), a target of TGF-b1, is needed for its short-term complex interacts with the FosB-gene chromatin and stimu- effects of TGF-b1 on cell motility involving Smad4- lates its transcription, and FosB in turn, utilizes FosB/histone dependent pathway (Michl et al.,2005). -

Analysis of Gene Expression Data for Gene Ontology
ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION A Thesis Presented to The Graduate Faculty of The University of Akron In Partial Fulfillment of the Requirements for the Degree Master of Science Robert Daniel Macholan May 2011 ANALYSIS OF GENE EXPRESSION DATA FOR GENE ONTOLOGY BASED PROTEIN FUNCTION PREDICTION Robert Daniel Macholan Thesis Approved: Accepted: _______________________________ _______________________________ Advisor Department Chair Dr. Zhong-Hui Duan Dr. Chien-Chung Chan _______________________________ _______________________________ Committee Member Dean of the College Dr. Chien-Chung Chan Dr. Chand K. Midha _______________________________ _______________________________ Committee Member Dean of the Graduate School Dr. Yingcai Xiao Dr. George R. Newkome _______________________________ Date ii ABSTRACT A tremendous increase in genomic data has encouraged biologists to turn to bioinformatics in order to assist in its interpretation and processing. One of the present challenges that need to be overcome in order to understand this data more completely is the development of a reliable method to accurately predict the function of a protein from its genomic information. This study focuses on developing an effective algorithm for protein function prediction. The algorithm is based on proteins that have similar expression patterns. The similarity of the expression data is determined using a novel measure, the slope matrix. The slope matrix introduces a normalized method for the comparison of expression levels throughout a proteome. The algorithm is tested using real microarray gene expression data. Their functions are characterized using gene ontology annotations. The results of the case study indicate the protein function prediction algorithm developed is comparable to the prediction algorithms that are based on the annotations of homologous proteins. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Hypertension and Prolonged Vasoconstrictor Signaling in RGS2- Deficient Mice
Amendment history: Addendum (April 2003) Hypertension and prolonged vasoconstrictor signaling in RGS2- deficient mice Scott P. Heximer, … , Robert P. Mecham, Kendall J. Blumer J Clin Invest. 2003;111(4):445-452. https://doi.org/10.1172/JCI15598. Article Cardiology Signaling by hormones and neurotransmitters that activate G protein–coupled receptors (GPCRs) maintains blood pressure within the normal range despite large changes in cardiac output that can occur within seconds. This implies that blood pressure regulation requires precise kinetic control of GPCR signaling. To test this hypothesis, we analyzed mice deficient in RGS2, a GTPase-activating protein that greatly accelerates the deactivation rate of heterotrimeric G proteins in vitro. Both rgs2+/– and rgs2–/– mice exhibited a strong hypertensive phenotype, renovascular abnormalities, persistent constriction of the resistance vasculature, and prolonged response of the vasculature to vasoconstrictors in vivo. Analysis of P2Y receptor–mediated Ca2+ signaling in vascular smooth muscle cells in vitro indicated that loss of RGS2 increased agonist potency and efficacy and slowed the kinetics of signal termination. These results establish that abnormally prolonged signaling by G protein–coupled vasoconstrictor receptors can contribute to the onset of hypertension, and they suggest that genetic defects affecting the function or expression of RGS2 may be novel risk factors for development of hypertension in humans. Find the latest version: https://jci.me/15598/pdf Hypertension and prolonged See the related Commentary beginning on page 441. vasoconstrictor signaling in RGS2-deficient mice Scott P. Heximer,1 Russell H. Knutsen,1 Xiaoguang Sun,1 Kevin M. Kaltenbronn,1 Man-Hee Rhee,1 Ning Peng,2 Antonio Oliveira-dos-Santos,3 Josef M. -

Regulation of Neuronal Gene Expression and Survival by Basal NMDA Receptor Activity: a Role for Histone Deacetylase 4
The Journal of Neuroscience, November 12, 2014 • 34(46):15327–15339 • 15327 Cellular/Molecular Regulation of Neuronal Gene Expression and Survival by Basal NMDA Receptor Activity: A Role for Histone Deacetylase 4 Yelin Chen,1 Yuanyuan Wang,1 Zora Modrusan,3 Morgan Sheng,1 and Joshua S. Kaminker1,2 Departments of 1Neuroscience, 2Bioinformatics and Computational Biology, and 3Molecular Biology, Genentech Inc., South San Francisco, California 94080 Neuronal gene expression is modulated by activity via calcium-permeable receptors such as NMDA receptors (NMDARs). While gene expression changes downstream of evoked NMDAR activity have been well studied, much less is known about gene expression changes that occur under conditions of basal neuronal activity. In mouse dissociated hippocampal neuronal cultures, we found that a broad NMDAR antagonist, AP5, induced robust gene expression changes under basal activity, but subtype-specific antagonists did not. While some of the gene expression changes are also known to be downstream of stimulated NMDAR activity, others appear specific to basal NMDARactivity.ThegenesalteredbyAP5treatmentofbasalcultureswereenrichedforpathwaysrelatedtoclassIIahistonedeacetylases (HDACs), apoptosis, and synapse-related signaling. Specifically, AP5 altered the expression of all three class IIa HDACs that are highly expressed in the brain, HDAC4, HDAC5, and HDAC9, and also induced nuclear accumulation of HDAC4. HDAC4 knockdown abolished a subset of the gene expression changes induced by AP5, and led to neuronal death under -

Micrornas in Cardiovascular Disease: from Pathogenesis to Prevention and Treatment
MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment Daniel Quiat, Eric N. Olson J Clin Invest. 2013;123(1):11-18. https://doi.org/10.1172/JCI62876. Review Series The management of cardiovascular risk through lifestyle modification and pharmacotherapy is paramount to the prevention of cardiovascular disease. Epidemiological studies have identified obesity, dyslipidemia, diabetes, and hypertension as interrelated factors that negatively affect cardiovascular health. Recently, genetic and pharmacological evidence in model systems has implicated microRNAs as dynamic modifiers of disease pathogenesis. An expanded understanding of the function of microRNAs in gene regulatory networks associated with cardiovascular risk will enable identification of novel genetic mechanisms of disease and inform the development of innovative therapeutic strategies. Find the latest version: https://jci.me/62876/pdf Review series MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment Daniel Quiat and Eric N. Olson Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, Texas, USA. The management of cardiovascular risk through lifestyle modification and pharmacotherapy is paramount to the prevention of cardiovascular disease. Epidemiological studies have identified obesity, dyslipidemia, diabetes, and hypertension as interrelated factors that negatively affect cardiovascular health. Recently, genetic and pharmaco- logical evidence in model systems has implicated microRNAs -

Spatial Distribution of Leading Pacemaker Sites in the Normal, Intact Rat Sinoa
Supplementary Material Supplementary Figure 1: Spatial distribution of leading pacemaker sites in the normal, intact rat sinoatrial 5 nodes (SAN) plotted along a normalized y-axis between the superior vena cava (SVC) and inferior vena 6 cava (IVC) and a scaled x-axis in millimeters (n = 8). Colors correspond to treatment condition (black: 7 baseline, blue: 100 µM Acetylcholine (ACh), red: 500 nM Isoproterenol (ISO)). 1 Supplementary Figure 2: Spatial distribution of leading pacemaker sites before and after surgical 3 separation of the rat SAN (n = 5). Top: Intact SAN preparations with leading pacemaker sites plotted during 4 baseline conditions. Bottom: Surgically cut SAN preparations with leading pacemaker sites plotted during 5 baseline conditions (black) and exposure to pharmacological stimulation (blue: 100 µM ACh, red: 500 nM 6 ISO). 2 a &DUGLDFIoQChDQQHOV .FQM FOXVWHU &DFQDG &DFQDK *MD &DFQJ .FQLS .FQG .FQK .FQM &DFQDF &DFQE .FQM í $WSD .FQD .FQM í .FQN &DVT 5\U .FQM &DFQJ &DFQDG ,WSU 6FQD &DFQDG .FQQ &DFQDJ &DFQDG .FQD .FQT 6FQD 3OQ 6FQD +FQ *MD ,WSU 6FQE +FQ *MG .FQN .FQQ .FQN .FQD .FQE .FQQ +FQ &DFQDD &DFQE &DOP .FQM .FQD .FQN .FQG .FQN &DOP 6FQD .FQD 6FQE 6FQD 6FQD ,WSU +FQ 6FQD 5\U 6FQD 6FQE 6FQD .FQQ .FQH 6FQD &DFQE 6FQE .FQM FOXVWHU V6$1 L6$1 5$ /$ 3 b &DUGLDFReFHSWRUV $GUDF FOXVWHU $GUDD &DY &KUQE &KUP &KJD 0\O 3GHG &KUQD $GUE $GUDG &KUQE 5JV í 9LS $GUDE 7SP í 5JV 7QQF 3GHE 0\K $GUE *QDL $QN $GUDD $QN $QN &KUP $GUDE $NDS $WSE 5DPS &KUP 0\O &KUQD 6UF &KUQH $GUE &KUQD FOXVWHU V6$1 L6$1 5$ /$ 4 c 1HXURQDOPURWHLQV -

Figure S1. HAEC ROS Production and ML090 NOX5-Inhibition
Figure S1. HAEC ROS production and ML090 NOX5-inhibition. (a) Extracellular H2O2 production in HAEC treated with ML090 at different concentrations and 24 h after being infected with GFP and NOX5-β adenoviruses (MOI 100). **p< 0.01, and ****p< 0.0001 vs control NOX5-β-infected cells (ML090, 0 nM). Results expressed as mean ± SEM. Fold increase vs GFP-infected cells with 0 nM of ML090. n= 6. (b) NOX5-β overexpression and DHE oxidation in HAEC. Representative images from three experiments are shown. Intracellular superoxide anion production of HAEC 24 h after infection with GFP and NOX5-β adenoviruses at different MOIs treated or not with ML090 (10 nM). MOI: Multiplicity of infection. Figure S2. Ontology analysis of HAEC infected with NOX5-β. Ontology analysis shows that the response to unfolded protein is the most relevant. Figure S3. UPR mRNA expression in heart of infarcted transgenic mice. n= 12-13. Results expressed as mean ± SEM. Table S1: Altered gene expression due to NOX5-β expression at 12 h (bold, highlighted in yellow). N12hvsG12h N18hvsG18h N24hvsG24h GeneName GeneDescription TranscriptID logFC p-value logFC p-value logFC p-value family with sequence similarity NM_052966 1.45 1.20E-17 2.44 3.27E-19 2.96 6.24E-21 FAM129A 129. member A DnaJ (Hsp40) homolog. NM_001130182 2.19 9.83E-20 2.94 2.90E-19 3.01 1.68E-19 DNAJA4 subfamily A. member 4 phorbol-12-myristate-13-acetate- NM_021127 0.93 1.84E-12 2.41 1.32E-17 2.69 1.43E-18 PMAIP1 induced protein 1 E2F7 E2F transcription factor 7 NM_203394 0.71 8.35E-11 2.20 2.21E-17 2.48 1.84E-18 DnaJ (Hsp40) homolog. -

PDF Hosted at the Radboud Repository of the Radboud University Nijmegen
PDF hosted at the Radboud Repository of the Radboud University Nijmegen The following full text is a publisher's version. For additional information about this publication click this link. http://hdl.handle.net/2066/147462 Please be advised that this information was generated on 2021-10-11 and may be subject to change. Chromatin Regulatory Complexes from an Interaction Proteomics Perspective an Interaction Proteomics from Regulatory Complexes Chromatin Chromatin Regulatory Complexes from an Interaction Proteomics Perspective H. İrem Baymaz H. İrem Baymaz H. İrem Baymaz 2015 Chromatin Regulatory Complexes from an Interaction Proteomics Perspective H. İrem Baymaz printed by: Proefschriftmaken.nl || Uitgeverij BOXPress Copyright © 2015 H. İrem Baymaz Chromatin Regulatory Complexes from an Interaction Proteomics Perspective Chromatine-Regulerende Complexen vanuit een Interactie-Proteomics Perspectief Proefschrift ter verkrijging van de graad van doctor aan de Radboud Universiteit Nijmegen op gezag van de rector magnificus prof. dr. Th.L.M. Engelen, volgens besluit van het college van decanen in het openbaar te verdedigen op donderdag 26 november 2015 om 12.30 uur precies door Hamdiye İrem Baymaz geboren op 8 maart 1986 te Ankara, Turkije Promoter: Prof. dr. M. Vermeulen Manuscriptcommissie: Prof. dr. J.H.L.M. van Bokhoven Prof. dr. H.T.M. Timmers (UMCU) Dr. L.M. Kamminga Anne ve Babama. Table of Contents List of Abbreviations 9 Chapter 1 11 Introduction Chapter 2 45 Identifying nuclear protein-protein interactions using GFP affinity purification -

Identification of Transcriptional Mechanisms Downstream of Nf1 Gene Defeciency in Malignant Peripheral Nerve Sheath Tumors Daochun Sun Wayne State University
Wayne State University DigitalCommons@WayneState Wayne State University Dissertations 1-1-2012 Identification of transcriptional mechanisms downstream of nf1 gene defeciency in malignant peripheral nerve sheath tumors Daochun Sun Wayne State University, Follow this and additional works at: http://digitalcommons.wayne.edu/oa_dissertations Recommended Citation Sun, Daochun, "Identification of transcriptional mechanisms downstream of nf1 gene defeciency in malignant peripheral nerve sheath tumors" (2012). Wayne State University Dissertations. Paper 558. This Open Access Dissertation is brought to you for free and open access by DigitalCommons@WayneState. It has been accepted for inclusion in Wayne State University Dissertations by an authorized administrator of DigitalCommons@WayneState. IDENTIFICATION OF TRANSCRIPTIONAL MECHANISMS DOWNSTREAM OF NF1 GENE DEFECIENCY IN MALIGNANT PERIPHERAL NERVE SHEATH TUMORS by DAOCHUN SUN DISSERTATION Submitted to the Graduate School of Wayne State University, Detroit, Michigan in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY 2012 MAJOR: MOLECULAR BIOLOGY AND GENETICS Approved by: _______________________________________ Advisor Date _______________________________________ _______________________________________ _______________________________________ © COPYRIGHT BY DAOCHUN SUN 2012 All Rights Reserved DEDICATION This work is dedicated to my parents and my wife Ze Zheng for their continuous support and understanding during the years of my education. I could not achieve my goal without them. ii ACKNOWLEDGMENTS I would like to express tremendous appreciation to my mentor, Dr. Michael Tainsky. His guidance and encouragement throughout this project made this dissertation come true. I would also like to thank my committee members, Dr. Raymond Mattingly and Dr. John Reiners Jr. for their sustained attention to this project during the monthly NF1 group meetings and committee meetings, Dr. -

I3C & Hormone Positive Breast Cancer
ARTICLE IN PRESS Journal of Nutritional Biochemistry xx (2006) xxx–xxx Estrogen receptor a as a target for indole-3-carbinol Thomas T.Y. Wanga,4, Matthew J. Milnerb, John A. Milnerc, Young S. Kimc aPhytonutrients Laboratory, Beltsville Human Nutrition Research Center, Agricultural Research Service, U.S. Department of Agriculture, Beltsville, MD 20705, USA bDepartment of Biology, The Pennsylvania State University, University Park, PA, USA cNutritional Science Research Group, Division of Cancer Prevention, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA Received 8 June 2005; received in revised form 14 October 2005; accepted 22 October 2005 Abstract A wealth of preclinical evidence supports the antitumorigenic properties of indole-3-carbinol (I3C), which is a major bioactive food component in cruciferous vegetables. However, the underlying molecular mechanism(s) accounting for these effects remain unresolved. In the present study, estrogen receptor alpha (ER-a) was identified as a potential molecular target for I3C. Treating MCF-7 cells with 100 AM I3C reduced ER-a mRNA expression by approximately 60% compared to controls. This reduction in ER-a transcript levels was confirmed using real-time polymerase chain reaction. The I3C dimer, 3,3V-diindolylmethane (DIM), was considerably more effective in depressing ER-a mRNA in MCF-7 cells than the monomeric unit. The suppressive effects of 5 AM DIM on ER-a mRNA was comparable to that caused by 100 AM I3C. DIM is known to accumulate in the nucleus and is a preferred ligand for aryl hydrocarbon receptor (AhR) to I3C. The addition of other AhR ligands, a-naphthoflavone (a-NF, 10 AM) and luteolin (10 AM), to the culture media resulted in a similar suppression in ER-a mRNA levels to that caused by 5 AM DIM. -
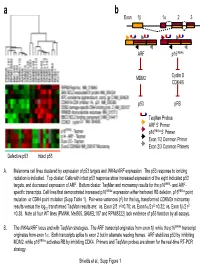
Taqman Probes ARF 5' Primer P16ink4a 5' Primer Exon 1/2
a b Exon 1β 1α 23 ARF p16INK4a MDM2 Cyclin D CDK4/6 p53 pRB TaqMan Probes ARF 5’ Primer p16INK4a 5’ Primer Exon 1/2 Common Primer Exon 2/3 Common Primers Defective p53 Intact p53 A. Melanoma cell lines clustered by expression of p53 targets and INK4a/ARF expression. The p53 response to ionizing radiation is indicated. Top cluster: Cells with intact p53 response show increased expression of the eight indicated p53 targets, and decreased expression of ARF. Bottom cluster: TaqMan and microarray results for the p16INK4- and ARF- specific transcripts. Cell lines that demonstrated increased p16INK4a expression either harbored RB deletion, p16INK4a point 2 mutation or CDK4 point mutation (Supp Table 1). Pair-wise variances (r ) for the log2 transformed CDKN2a microarray 2 2 2 results versus the log10 transformed TaqMan results are: vs. Exon 2/3 r =0.78; vs. Exon1α/2 r =0.32; vs. Exon 1β/2 r =0.38. Note all four WT lines (PMWK, Mel505, SKMEL187 and RPMI8322) lack evidence of p53 function by all assays. B. The INK4a/ARF locus and with TaqMan strategies. The ARF transcript originates from exon 1β while the p16INK4a transcript originates from exon 1α. Both transcripts splice to exon 2 but in alternate reading frames. ARF stabilizes p53 by inhibiting MDM2, while p16INK4a activates RB by inhibiting CDK4. Primers and TaqMan probes are shown for the real-time RT-PCR strategy. Shields et al., Supp Figure 1 SKMEL 28 U01 24h SKMEL WM2664 U01 48h WM2664 U01 24h 24 U01 48h SKMEL 24 U01 24h SKMEL 24 Untreated SKMEL 24 DMSO 48h SKMEL 24 DMSO 24h SKMEL WM2664 DMSO 24h WM2664 DMSO 48h WM2644 Untreated 28 Untreated SKMEL 28 DMSO 48h SKMEL 28 DMSO 24h SKMEL -3.00 -2.00 -1.00 0.00 1.00 2.00 3.00 relative to median expression Genes decreased by UO1 (863) HIF1A Hypoxia-inducible factor 1, alpha subunit NM_001530 RBBP8 Retinoblastoma binding protein 8 NM_002894 Homo sapiens, clone IMAGE:4337652, mRNA BC018676 EIF4EBP1 Eukaryotic translation initiation factor 4E binding protein EXOSC8 Exosome component 8 NM_181503 ENST00000321524 MCM7 MCM7 minichromosome maintenance deficient 7 (S.