Altered Myeloid & Lymphoid Composition of Tumor
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Semaphorin 4D Promotes Skeletal Metastasis in Breast Cancer
UCLA UCLA Previously Published Works Title Semaphorin 4D Promotes Skeletal Metastasis in Breast Cancer. Permalink https://escholarship.org/uc/item/1vj106ww Journal PloS one, 11(2) ISSN 1932-6203 Authors Yang, Ying-Hua Buhamrah, Asma Schneider, Abraham et al. Publication Date 2016 DOI 10.1371/journal.pone.0150151 Peer reviewed eScholarship.org Powered by the California Digital Library University of California RESEARCH ARTICLE Semaphorin 4D Promotes Skeletal Metastasis in Breast Cancer Ying-Hua Yang1, Asma Buhamrah1, Abraham Schneider1,2, Yi-Ling Lin3, Hua Zhou1, Amr Bugshan1, John R. Basile1,2* 1 Department of Oncology and Diagnostic Sciences, University of Maryland Dental School, Baltimore, Maryland, United States of America, 2 Greenebaum Cancer Center, Baltimore, Maryland, United States of America, 3 Division of Diagnostic and Surgical Sciences, School of Dentistry, University of California Los Angeles, Los Angeles, California, United States of America * [email protected] Abstract Bone density is controlled by interactions between osteoclasts, which resorb bone, and osteoblasts, which deposit it. The semaphorins and their receptors, the plexins, originally shown to function in the immune system and to provide chemotactic cues for axon guid- ance, are now known to play a role in this process as well. Emerging data have identified OPEN ACCESS Semaphorin 4D (Sema4D) as a product of osteoclasts acting through its receptor Plexin-B1 on osteoblasts to inhibit their function, tipping the balance of bone homeostasis in favor of Citation: Yang Y-H, Buhamrah A, Schneider A, Lin Y- resorption. Breast cancers and other epithelial malignancies overexpress Sema4D, so we L, Zhou H, Bugshan A, et al. (2016) Semaphorin 4D Promotes Skeletal Metastasis in Breast Cancer. -

Retinoid X Receptor Suppresses a Metastasis-Promoting Transcriptional Program in Myeloid Cells Via a Ligand-Insensitive Mechanism
Retinoid X receptor suppresses a metastasis-promoting transcriptional program in myeloid cells via a ligand-insensitive mechanism Mate Kissa, Zsolt Czimmerera,b, Gergely Nagyb, Pawel Bieniasz-Krzywiecc,d, Manuel Ehlingc,d, Attila Papa, Szilard Poliskaa,e, Pal Botof, Petros Tzerpose, Attila Horvatha, Zsuzsanna Kolostyaka, Bence Danielg, Istvan Szatmarif, Massimiliano Mazzonec,d, and Laszlo Nagya,b,g,1 aNuclear Receptor Research Laboratory, Department of Biochemistry and Molecular Biology, University of Debrecen, 4032 Debrecen, Hungary; bMTA-DE “Lendület” Immunogenomics Research Group, University of Debrecen, 4032 Debrecen, Hungary; cLaboratory of Tumor Inflammation and Angiogenesis, Center for Cancer Biology, VIB, 3000 Leuven, Belgium; dLaboratory of Tumor Inflammation and Angiogenesis, Department of Oncology, KU Leuven, 3000 Leuven, Belgium; eUD-GenoMed Ltd., 4032 Debrecen, Hungary; fStem Cell Differentiation Laboratory, Department of Biochemistry and Molecular Biology, University of Debrecen, 4032 Debrecen, Hungary; and gGenomic Control of Metabolism Program, Sanford Burnham Prebys Medical Discovery Institute at Lake Nona, Orlando, FL 32827 Edited by Bert W. O’Malley, Baylor College of Medicine, Houston, TX, and approved August 18, 2017 (received for review January 18, 2017) Retinoid X receptor (RXR) regulates several key functions in myeloid and identified a network of RXR-bound enhancers that control cells, including inflammatory responses, phagocytosis, chemokine angiogenic genes including Vegfa, Hbegf, Litaf,andHipk2 (9). secretion, and proangiogenic activity. Its importance, however, in Activation of these enhancers by RXR induced a proangiogenic tumor-associated myeloid cells is unknown. In this study, we transcriptional program and phenotype (9). Interestingly, we demonstrate that deletion of RXR in myeloid cells enhances lung found that half of the 5,200 RXR-occupied genomic sites were metastasis formation while not affecting primary tumor growth. -

Oestrogen Receptor α AF-1 and AF-2 Domains Have Cell
ARTICLE DOI: 10.1038/s41467-018-07175-0 OPEN Oestrogen receptor α AF-1 and AF-2 domains have cell population-specific functions in the mammary epithelium Stéphanie Cagnet1, Dalya Ataca 1, George Sflomos1, Patrick Aouad1, Sonia Schuepbach-Mallepell2, Henry Hugues3, Andrée Krust4, Ayyakkannu Ayyanan1, Valentina Scabia1 & Cathrin Brisken 1 α α 1234567890():,; Oestrogen receptor (ER ) is a transcription factor with ligand-independent and ligand- dependent activation functions (AF)-1 and -2. Oestrogens control postnatal mammary gland development acting on a subset of mammary epithelial cells (MECs), termed sensor cells, which are ERα-positive by immunohistochemistry (IHC) and secrete paracrine factors, which stimulate ERα-negative responder cells. Here we show that deletion of AF-1 or AF-2 blocks pubertal ductal growth and subsequent development because both are required for expres- sion of essential paracrine mediators. Thirty percent of the luminal cells are ERα-negative by IHC but express Esr1 transcripts. This low level ERα expression through AF-2 is essential for cell expansion during puberty and growth-inhibitory during pregnancy. Cell-intrinsic ERα is not required for cell proliferation nor for secretory differentiation but controls transcript levels of cell motility and cell adhesion genes and a stem cell and epithelial mesenchymal transition (EMT) signature identifying ERα as a key regulator of mammary epithelial cell plasticity. 1 Swiss Institute for Experimental Cancer Research, School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, CH-1015 Lausanne, Switzerland. 2 Department of Biochemistry, University of Lausanne, CH-1066 Epalinges, Switzerland. 3 Centre Hospitalier Universitaire Vaudois, Department of Laboratory Medecine, University Hospital of Lausanne, CH-1011 Lausanne, Switzerland. -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int. -

Molecular Signatures Differentiate Immune States in Type 1 Diabetes Families
Page 1 of 65 Diabetes Molecular signatures differentiate immune states in Type 1 diabetes families Yi-Guang Chen1, Susanne M. Cabrera1, Shuang Jia1, Mary L. Kaldunski1, Joanna Kramer1, Sami Cheong2, Rhonda Geoffrey1, Mark F. Roethle1, Jeffrey E. Woodliff3, Carla J. Greenbaum4, Xujing Wang5, and Martin J. Hessner1 1The Max McGee National Research Center for Juvenile Diabetes, Children's Research Institute of Children's Hospital of Wisconsin, and Department of Pediatrics at the Medical College of Wisconsin Milwaukee, WI 53226, USA. 2The Department of Mathematical Sciences, University of Wisconsin-Milwaukee, Milwaukee, WI 53211, USA. 3Flow Cytometry & Cell Separation Facility, Bindley Bioscience Center, Purdue University, West Lafayette, IN 47907, USA. 4Diabetes Research Program, Benaroya Research Institute, Seattle, WA, 98101, USA. 5Systems Biology Center, the National Heart, Lung, and Blood Institute, the National Institutes of Health, Bethesda, MD 20824, USA. Corresponding author: Martin J. Hessner, Ph.D., The Department of Pediatrics, The Medical College of Wisconsin, Milwaukee, WI 53226, USA Tel: 011-1-414-955-4496; Fax: 011-1-414-955-6663; E-mail: [email protected]. Running title: Innate Inflammation in T1D Families Word count: 3999 Number of Tables: 1 Number of Figures: 7 1 For Peer Review Only Diabetes Publish Ahead of Print, published online April 23, 2014 Diabetes Page 2 of 65 ABSTRACT Mechanisms associated with Type 1 diabetes (T1D) development remain incompletely defined. Employing a sensitive array-based bioassay where patient plasma is used to induce transcriptional responses in healthy leukocytes, we previously reported disease-specific, partially IL-1 dependent, signatures associated with pre and recent onset (RO) T1D relative to unrelated healthy controls (uHC). -

NK Cells: Receptors and Functions Eric Vivier and Sophie Ugolini
NK cells: receptors and functions Eric Vivier and Sophie Ugolini Natural killer (NK) cells were identified in 1975 as lymphocytes of the innate many processes that were originally restricted to adaptive immunity, such immune system that can kill tumour cells. Since then, NK cells have been as priming, education and memory, are now known to occur in NK cells. shown to kill an array of ‘stressed’ cells and secrete cytokines that Indeed, NK cells undergo sophisticated processes of adaptation that allow participate in shaping adaptive immune responses. A key feature of NK cells them to be tuned to their environment. There is also a growing interest in resides in their capacity to distinguish stressed cells (such as tumour cells, manipulating NK cells in innovative therapeutic settings. For example, the infected cells and damaged cells) from normal cells. Although NK cells are understanding of NK cell inhibition by MHC class I-specific receptors has generally considered to be components of early innate immune defence, prompted the design of innovative anticancer therapies. The NK cell detection system includes numerous receptors, Key receptors on human NK cells the engagement of which dictates the quality and intensity Inhibitory receptors Activating receptors, adhesion molecules or co-stimulatory molecules of the NK cell response. NK cells use inhibitory receptors to gauge the absence of constitutively expressed self molecules Target cell Nectin 2 SLAMF7 Sialic CLEC2D (CD112, PVRL2) NECL2 L-selectin NECL5 AICL NTB-A (CRACC, on susceptible target cells. As a consequence, NK cells can Receptors for MHC class I and class I-Iike molecules B7-H6 acid Cadherins (LLT1) CMV pp65 NECL5 (CADM1) (CD62L) (CD155, PVR) (CLEC2B) CEACAM1 (SLAMF6) CS1, CD319) recognize ‘missing self’ on haematopoietic cells. -

Supplemental Figure Legends
Supplemental Figure Legends Supplemental Figure 1. Expression of cytoplasmic immunoglobulin in D4 cell subsets. A. Phenotype of D4 cell subsets. B. Expression of cyIgM, cyIgA, and cyIgG in D4 cell subsets. Results are those of one experiment representative of three, SI means Staining Index. C. Expression of cyIgM, cyIgA, and cyIgG in D4 cell subsets. Results are mean SI obtained in three separate experiments. * The mean expression is significantly different from that in CD20high B cells. ** The mean expression is significantly different from that in D4 CD20low prePBs. Supplemental Figure 2. Expression of genes coding for B or PC markers. Data are the Affymetrix signals of expression of genes coding for B or PC markers in MBCs, CD20high B cells, CD20low prePBs, CD20- prePBs, PBs, early PCs, and BMPCs using the same color code as in Figure 6A. Data are the mean value ± SD of gene expression determined in 5 separate experiments. * The mean expression is significantly different from that in CD20high B cells. ** The mean expression is significantly different from that in D4 CD20low prePBs. Supplemental Figure 3. Expression of genes coding for homing molecules. Data are the Affymetrix signals of expression of genes coding for remarkable chemokine receptors or integrins in MBCs, CD20high B cells, CD20low prePBs, CD20- prePBs, PBs, early PCs, and BMPCs using the same color code as in Figure 6A. Data are the mean value ± SD of gene expression determined in 5 separate experiments. * The mean expression is significantly different from that in CD20high B cells. ** The mean expression is significantly different from that in D4 CD20low prePBs. -

Interleukins As Mediators of the Tumor Cell—Bone Cell Crosstalk During the Initiation of Breast Cancer Bone Metastasis
International Journal of Molecular Sciences Review Interleukins as Mediators of the Tumor Cell—Bone Cell Crosstalk during the Initiation of Breast Cancer Bone Metastasis Marie-Therese Haider 1, Nicole Ridlmaier 1,2, Daniel J. Smit 3 and Hanna Taipaleenmäki 1,* 1 Molecular Skeletal Biology Laboratory, Department of Trauma and Orthopedic Surgery, University Medical Center Hamburg-Eppendorf, 20246 Hamburg, Germany; [email protected] (M.-T.H.); [email protected] (N.R.) 2 Department of Life Sciences, IMC FH Krems University of Applied Sciences, 3500 Krems, Austria 3 Institute of Biochemistry and Signal Transduction, University Medical Center Hamburg-Eppendorf, 20246 Hamburg, Germany; [email protected] * Correspondence: [email protected] Abstract: Patients with advanced breast cancer are at high risk of developing bone metastasis. Despite treatment advances for primary breast cancer, metastatic bone disease remains incurable with a low relative survival. Hence, new therapeutic approaches are required to improve survival and treatment outcome for these patients. Bone is among the most frequent sites of metastasis in breast cancer. Once in the bone, disseminated tumor cells can acquire a dormant state and remain quiescent until they resume growth, resulting in overt metastasis. At this stage the disease is characterized by excessive, osteoclast-mediated osteolysis. Cells of the bone microenvironment including osteoclasts, osteoblasts and endothelial cells contribute to the initiation and progression of breast cancer bone metastasis. Direct cell-to-cell contact as well as soluble factors regulate the crosstalk between disseminated breast cancer cells and bone cells. In this complex signaling network Citation: Haider, M.-T.; Ridlmaier, interleukins (ILs) have been identified as key regulators since both, cancer cells and bone cells secrete N.; Smit, D.J.; Taipaleenmäki, H. -

SUPPLEMENTAL FIGURE LEGEND Supplemental Figure 1. Representative HGA Histology of (A) Greater Than Median, and (B) Less Than
SUPPLEMENTAL FIGURE LEGEND Supplemental figure 1. Representative HGA histology of (A) greater than median, and (B) less than median tumor infiltration of helper T-cells (CD4; brown), (C) greater than median, and (D) less than median tumor infiltration of cytotoxic T-cells (CD8; red), (E) greater than 75th percentile, and (F) less than 75% percentile tumor infiltration of microglia/macrophages (AIF1; brown). Immunohistochemistry was performed using FFPE tumor sections and with hematoxylin counterstaining (400x magnification). Supplemental table IA. High grade astrocytoma long-term survivor patient details. Sample Survival Dead Age Karnofsky/ Dx detail location of Extent of Therapy received Used in ID (yrs) at Lansky tumor surgery microarray Dx performance study (yrs) score HGA1 17.0 N 34 100 AA pons biopsy radiation and BCNU HGA2 16.5 N 8 80 AA cerebrum GTR carboplatin, etoposide, CCNU, vincristine and radiation HGA3 16.0 N 47 90 GBM 1o cerebrum STR radiation and temozolmide HGA4 11.0 N 45 60 GBM, giant cell cerebrum GTR none HGA5 10.5 N 27 100 GBM 1o, cerebrum GTR radiation, CCNU, irinotecan, and temozolmide epithelioid HGA6 8.5 Y 21 100 RIG cerebellum STR radiation and temozolmide HGA7 8.5 N 2 80 GBM 1o thalamus/ STR cisplatin, vincristine, cytoxan, etoposide, carboplatin, cerebrum thiotepa HGA8 8.5 N 42 100 GBM 2o cerebrum GTR radiation, CCNU, irinotecan and temozolmide HGA9 8.5 N 46 80 GBM 1o cerebrum GTR radiation, CCNU, erlotinib and temozolmide yes HGA10 7.0 N 10 80 GBM 1o thalamus biopsy radiation and temozolmide HGA11 7.0 Y 24 -
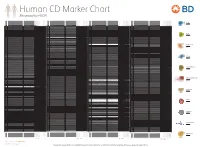
Reviewed by HLDA1
Human CD Marker Chart Reviewed by HLDA1 T Cell Key Markers CD3 CD4 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD8 CD1a R4, T6, Leu6, HTA1 b-2-Microglobulin, CD74 + + + – + – – – CD74 DHLAG, HLADG, Ia-g, li, invariant chain HLA-DR, CD44 + + + + + + CD158g KIR2DS5 + + CD248 TEM1, Endosialin, CD164L1, MGC119478, MGC119479 Collagen I/IV Fibronectin + ST6GAL1, MGC48859, SIAT1, ST6GALL, ST6N, ST6 b-Galactosamide a-2,6-sialyl- CD1b R1, T6m Leu6 b-2-Microglobulin + + + – + – – – CD75 CD22 CD158h KIR2DS1, p50.1 HLA-C + + CD249 APA, gp160, EAP, ENPEP + + tranferase, Sialo-masked lactosamine, Carbohydrate of a2,6 sialyltransferase + – – + – – + – – CD1c M241, R7, T6, Leu6, BDCA1 b-2-Microglobulin + + + – + – – – CD75S a2,6 Sialylated lactosamine CD22 (proposed) + + – – + + – + + + CD158i KIR2DS4, p50.3 HLA-C + – + CD252 TNFSF4, -

International Immunopharmacology 67 (2019) 220–230
International Immunopharmacology 67 (2019) 220–230 Contents lists available at ScienceDirect International Immunopharmacology journal homepage: www.elsevier.com/locate/intimp Semaphoring 4D is required for the induction of antioxidant stress and anti- inflammatory effects of dihydromyricetin in colon cancer T ⁎ Jun Lianga,1, Jing Wub,1, Fei Wangb, Pengfei Zhangb, Xuemei Zhangb, a Oncology Medicine Center, Shanghai East Hospital, Tongji University School of Medicine, Shanghai 200120, China b Department of Oncology, Shanghai East Hospital, Tongji University School of Medicine, Shanghai 200120, China ARTICLE INFO ABSTRACT Keywords: Semaphorin 4D (Sema4D) has been involved in cancer progression, the expression of which is associated with Dihydromyricetin the poor clinical outcomes of some cancer patients. Dihydromyricetin (DMY) has antitumor potentials for dif- Colorectal cancer ferent types of human cancer cells. However, the pharmacological effects of DMY on colon cancer (CC) or the Semaphorin 4D regulatory effects of Sema4D on this process remain largely unknown. In the present study, we aimed to evaluate Oxidative stress the effects of DMY on CC, and to elucidate the role of Sema4D in DMY-induced antitumor effects. DMY inhibited Inflammation the proliferation and growth of Colo-205 colon cancer cells significantly in vivo and in vitro. DMY inhibited reactive oxygen species (ROS) and malondialdehyde (MDA) levels, but increased glutathione (GSH) level. Moreover, the activities of antioxidant enzymes catalase (CAT), superoxide dismutase (SOD), glutathione per- oxidase (GPx), glutathione reductase (GR) and heme oxygenase 1 (HO-1) were enhanced by DMY treatment in vitro, showing strong anti-oxidative stress effect. In addition, DMY inhibited the secretion of interleukin 1β (IL- 1β), interleukin-6 (IL-6), interleukin-8 (IL-8) and tumor necrosis factor (TNF-α) in the supernatant of Colo-205 culture medium.