Evidence for Strain-Specific Exometabolomic Responses of The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

University of Oklahoma
UNIVERSITY OF OKLAHOMA GRADUATE COLLEGE MACRONUTRIENTS SHAPE MICROBIAL COMMUNITIES, GENE EXPRESSION AND PROTEIN EVOLUTION A DISSERTATION SUBMITTED TO THE GRADUATE FACULTY in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY By JOSHUA THOMAS COOPER Norman, Oklahoma 2017 MACRONUTRIENTS SHAPE MICROBIAL COMMUNITIES, GENE EXPRESSION AND PROTEIN EVOLUTION A DISSERTATION APPROVED FOR THE DEPARTMENT OF MICROBIOLOGY AND PLANT BIOLOGY BY ______________________________ Dr. Boris Wawrik, Chair ______________________________ Dr. J. Phil Gibson ______________________________ Dr. Anne K. Dunn ______________________________ Dr. John Paul Masly ______________________________ Dr. K. David Hambright ii © Copyright by JOSHUA THOMAS COOPER 2017 All Rights Reserved. iii Acknowledgments I would like to thank my two advisors Dr. Boris Wawrik and Dr. J. Phil Gibson for helping me become a better scientist and better educator. I would also like to thank my committee members Dr. Anne K. Dunn, Dr. K. David Hambright, and Dr. J.P. Masly for providing valuable inputs that lead me to carefully consider my research questions. I would also like to thank Dr. J.P. Masly for the opportunity to coauthor a book chapter on the speciation of diatoms. It is still such a privilege that you believed in me and my crazy diatom ideas to form a concise chapter in addition to learn your style of writing has been a benefit to my professional development. I’m also thankful for my first undergraduate research mentor, Dr. Miriam Steinitz-Kannan, now retired from Northern Kentucky University, who was the first to show the amazing wonders of pond scum. Who knew that studying diatoms and algae as an undergraduate would lead me all the way to a Ph.D. -

"Plastid Originand Evolution". In: Encyclopedia of Life
CORE Metadata, citation and similar papers at core.ac.uk Provided by University of Queensland eSpace Plastid Origin and Advanced article Evolution Article Contents . Introduction Cheong Xin Chan, Rutgers University, New Brunswick, New Jersey, USA . Primary Plastids and Endosymbiosis . Secondary (and Tertiary) Plastids Debashish Bhattacharya, Rutgers University, New Brunswick, New Jersey, USA . Nonphotosynthetic Plastids . Plastid Theft . Plastid Origin and Eukaryote Evolution . Concluding Remarks Online posting date: 15th November 2011 Plastids (or chloroplasts in plants) are organelles within organisms that emerged ca. 2.8 billion years ago (Olson, which photosynthesis takes place in eukaryotes. The ori- 2006), followed by the evolution of eukaryotic algae ca. 1.5 gin of the widespread plastid traces back to a cyano- billion years ago (Yoon et al., 2004) and finally by the rise of bacterium that was engulfed and retained by a plants ca. 500 million years ago (Taylor, 1988). Photosynthetic reactions occur within the cytosol in heterotrophic protist through a process termed primary prokaryotes. In eukaryotes, however, the reaction takes endosymbiosis. Subsequent (serial) events of endo- place in the organelle, plastid (e.g. chloroplast in plants). symbiosis, involving red and green algae and potentially The plastid also houses many other reactions that are other eukaryotes, yielded the so-called ‘complex’ plastids essential for growth and development in algae and plants; found in photosynthetic taxa such as diatoms, dino- for example, the -

Influence of the Calcium Carbonate Shell Of
fmars-08-664269 June 30, 2021 Time: 12:15 # 1 ORIGINAL RESEARCH published: 30 June 2021 doi: 10.3389/fmars.2021.664269 Influence of the Calcium Carbonate Shell of Coccolithophores on Ingestion and Growth of a Dinoflagellate Predator Mathias Haunost1*, Ulf Riebesell1, Francesco D’Amore1, Ole Kelting1 and Lennart T. Bach2 1 GEOMAR Helmholtz Centre for Ocean Research Kiel, Kiel, Germany, 2 Institute for Marine and Antarctic Studies, Ecology & Biodiversity, University of Tasmania, Hobart, TAS, Australia Coccolithophores are an important group of ∼200 marine phytoplankton species which cover themselves with a calcium carbonate shell called “coccosphere.” Coccolithophores are ecologically and biogeochemically important but the reason why they calcify remains elusive. One key function may be that the coccosphere offers protection against microzooplankton predation, which is one of the main causes of phytoplankton death in the ocean. Here, we investigated the effect of the coccosphere on ingestion and growth of the heterotrophic dinoflagellate Oxyrrhis marina. Calcified and decalcified cells of the coccolithophore species Emiliania huxleyi, Pleurochrysis carterae, and Gephyrocapsa oceanica were offered separately to the predator as well ∼ Edited by: as in an initial 1:1 mixture. The decrease of the prey concentrations and predator Per Juel Hansen, abundances were monitored over a period of 48–72 h. We found that O. marina did University of Copenhagen, Denmark not actively select against calcified cells, but rather showed a size selective feeding Reviewed by: behavior. Thus, the coccosphere does not provide a direct protection against grazing by Jozef I. Nissimov, University of Waterloo, Canada O. marina. However, O. marina showed slower growth when calcified coccolithophores Rong Bi, were fed. -

Selective Algicidal Action of Peptides Against Harmful Algal Bloom Species
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central Selective Algicidal Action of Peptides against Harmful Algal Bloom Species Seong-Cheol Park1, Jong-Kook Lee1, Si Wouk Kim2, Yoonkyung Park1,3* 1 Research Center for Proteinaceous Materials (RCPM), Chosun University, Gwangju, Republic of Korea, 2 Department of Environmental Engineering, Chosun University, Gwangju, Republic of Korea, 3 Department of Biotechnology, Chosun University, Gwangju, Republic of Korea Abstract Recently, harmful algal bloom (HAB), also termed ‘‘red tide’’, has been recognized as a serious problem in marine environments according to climate changes worldwide. Many novel materials or methods to prevent HAB have not yet been employed except for clay dispersion, in which can the resulting sedimentation on the seafloor can also cause alteration in marine ecology or secondary environmental pollution. In the current study, we investigated that antimicrobial peptide have a potential in controlling HAB without cytotoxicity to harmless marine organisms. Here, antimicrobial peptides are proposed as new algicidal compounds in combating HAB cells. HPA3 and HPA3NT3 peptides which exert potent antimicrobial activity via pore forming action in plasma membrane showed that HPA3NT3 reduced the motility of algal cells, disrupted their plasma membrane, and induced the efflux of intracellular components. Against raphidoflagellate such as Heterosigma akashiwo, Chattonella sp., and C. marina, it displayed a rapid lysing action in cell membranes at 1,4 mM within 2 min. Comparatively, its lysing effects occurred at 8 mM within 1 h in dinoflagellate such as Cochlodium polykrikoides, Prorocentrum micans, and P. minimum. Moreover, its lysing action induced the lysis of chloroplasts and loss of chlorophyll a. -
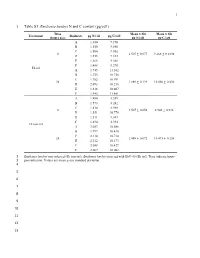
1 Table S1. Emiliania Huxleyi N and C Content (Pg/Cell)
1 1 Table S1. Emiliania huxleyi N and C content (pg/cell) Time Mean ± SD Mean ± SD Treatment Replicate pg N/cell pg C/cell (hours p.i.) pg N/cell pg C/cell A 1.580 9.190 B 1.550 9.580 C 1.500 9.036 0 1.507 ± 0.077 9.268 ± 0.1898 D 1.555 9.165 E 1.366 9.368 F 1.488 9.270 Eh inf A 1.945 11.102 B 1.755 10.758 C 1.782 10.991 24 1.889 ± 0.129 10.856 ± 0.430 D 2.093 10.216 E 1.816 10.607 F 1.945 11.461 A 1.480 8.559 B 1.573 8.242 C 1.434 8.988 0 1.507 ± 0.050 8.940 ± 0.936 D 1.551 10.778 E 1.511 8.681 F 1.494 8.394 Eh non-inf A 2.053 10.686 B 1.997 10.410 C 2.110 10.714 24 2.088 ± 0.072 10.415 ± 0.258 D 2.212 10.173 E 2.088 10.427 F 2.067 10.082 2 Emiliania huxleyi non-infected (Eh non-inf); Emiliania huxleyi infected with EhV-86 (Eh inf). Time indicate hours 3 post-infection. Values are mean ± one standard deviation. 4 5 6 7 8 9 10 11 12 13 O. MARINA RESPONSE TO INFECTED PREY 2 14 Table S2. Oxyrrhis marina’s average growth (day-1) and grazing rates (Eh cells Om-1 h-1 15 (experiment1) or Eh cells Om-1 day-1 (experiments 2 – 4) for each experiment under both diet 16 conditions. -

Feeding by Common Heterotrophic Protist Predators on Seven Prorocentrum Species
Research Article Algae 2020, 35(1): 61-78 https://doi.org/10.4490/algae.2020.35.2.28 Open Access Feeding by common heterotrophic protist predators on seven Prorocentrum species Ji Hyun You1, Hae Jin Jeong1,2,*, Hee Chang Kang1, Jin Hee Ok1, Sang Ah Park1 and An Suk Lim3 1School of Earth and Environmental Sciences, College of Natural Sciences, Seoul National University, Seoul 08826, Korea 2Research Institute of Oceanography, Seoul National University, Seoul 08826, Korea 3Division of Life Science, Gyeongsang National University, Jinju 52828, Korea Species belonging to the dinoflagellate genus Prorocentrum are known to cause red tides or harmful algal blooms. To understand the dynamics of a Prorocentrum sp., its growth and mortality due to predation need to be assessed. However, there are only a few Prorocentrum spp. for which heterotrophic protist predators have been reported. We explored feed- ing by the common heterotrophic dinoflagellates Gyrodinium dominans, Oxyrrhis marina, Pfiesteria piscicida, Oblea rotunda, and Polykrikos kofoidii and the naked ciliate Strombidinopsis sp. (approx. 90 µm cell length) on the planktonic species Prorocentrum triestinum, P. cordatum, P. donghaiense, P. rhathymum, and P. micans as well as the benthic spe- cies P. lima and P. hoffmannianum. All heterotrophic protists tested were able to feed on the planktonic prey species. However, O. marina and O. rotunda did not feed on P. lima and P. hoffmannianum, while G. dominans, P. kofoidii, and Strombidinopsis sp. did. The growth and ingestion rates of G. dominans and P. kofoidii on one of the seven Prorocentrum spp. were significantly different from those on other prey species. -

Growth Characteristics of Oxyrrhis Marina and Chattonella Marina in Their Co-Culture Systems
Nature Environment and Pollution Technology ISSN: 0972-6268 Vol. 14 No. 3 pp. 553-556 2015 An International Quarterly Scientific Journal Original Research Paper Growth Characteristics of Oxyrrhis marina and Chattonella marina in their Co-culture Systems Xinlong An, Xuemei Li and Zhixia Li Ocean College, Agricultural University of Hebei, Qinhuangdao 066003, Hebei, China ABSTRACT Nat. Env. & Poll. Tech. Website: www.neptjournal.com This study was aimed to investigate the growth characteristics of Oxyrrhis marina and Chattonella marina in co-culture to provide experimental evidences for discussing successions of harmful algal blooms (HABs) Received: 10-5-2015 and coastal biological communities. The colour changes of culture media of C. marina and growth Accepted: 21-6-2015 characteristics of O. marina and C. marina in co-culture were analysed by the combined methods of macro- Key Words: observation, microscopic examination and counting. In co-culture, the colours of culture media of C. marina Oxyrrhis marina had changed and their transparencies had increased with increasing elapsed incubation time after inoculated Chattonella marina by O. marina under different initial cell densities. With the increase of the initial density of O. marina (0.17×104 Growth characteristics cells/mL, 0.50×104 cells/mL and 0.64×104 cells/mL in C. marina culture media), the time required, that the Co-culture populations of O. marina reached the stationary phases, was shorter i.e. 6d, 5d and 3d after inoculated by O. marina, respectively, and the death time of all cells of C. marina became shorter, i.e. 7d, 6d and 4d after inoculated by O. -

A Winning Strategy of the Planktonic Flagellate Prymnesium Parvum
AQUATIC MICROBIAL ECOLOGY Vol. 32: 73–84, 2003 Published May 12 Aquat Microb Ecol Kill and eat your predator: a winning strategy of the planktonic flagellate Prymnesium parvum Urban Tillmann* Alfred Wegener Institute, Am Handelshafen 12, 27570 Bremerhaven, Germany ABSTRACT: Interactions between the toxic and mixotrophic haptophyte Prymnesium parvum and the heterotrophic dinoflagellate Oxyrrhis marina were investigated in P-limited semi-continuous cultures and nutrient-replete batch culture experiments. When exposed to 100 × 103 cells ml–1 of P- limited P. parvum, starved O. marina initially ingested the algae, but ingestion was low (0.07 cells grazer–1 h–1) compared to the O. marina ingestion rate when the Cryptophyte Rhodomonas sp. was offered as food (2.75 cells grazer–1 h–1). Microscopic observations showed that low ingestion is due to toxic effects of P. parvum on the dinoflagellate. Cells of O. marina lost their normal cell shape and became rounded, hyaline and finally lysed. Rounded and partly lysed O. marina cells were rapidly attacked by several P. parvum cells, which formed larger aggregates around the remains of the O. marina cells. This was accompanied by phagotrophic ingestion of particulate material originating from disintegrating O. marina. Cell-free culture medium lysed O. marina cells, although to a lower degree compared to the effect of algal suspensions. O. marina mortality was not only reduced by diluting P. parvum, but also by increasing the dinoflagellate concentration. This clearly indicates that the toxin is removed from the system by its action, presumably by binding to the membrane. Under conditions where toxic effects were not apparent (nutrient-replete batch cultures, low cell concentra- tions, dim light), P. -

The Israeli Journal of Aquaculture - Bamidgeh, IJA 69.2017.1385, 19 Pages
The Israeli Journal of Aquaculture - Bamidgeh, IJA_69.2017.1385, 19 pages The IJA appears exclusively as a peer-reviewed on-line open-access journal at http://www.siamb.org.il. To read papers free of charge, please register online at registration form. Sale of IJA papers is strictly forbidden. Development of Polyculture and Integrated Multi - Trophic Aquaculture (IMTA) in Israel: A Review Amir Neori1,2*, Muki Shpigel1, Lior Guttman1, Alvaro Israel3 1 National Center for Mariculture, Israel Oceanographic & Limnological Research, Eilat 8811201, Israel 2 Helmsley Charitable Trust Mediterranean Sea Research Center, Sedot Yam, The Leon H. Charney School of Marine Sciences, University of Haifa, Israel 3 The National Institute of Oceanography, Israel Oceanographic & Limnological Research, Haifa 3108001, Israel, and Spanish Bank of Algae, Universidad de Las Palmas de Gran Canaria, Canary Islands, Spain Key words: fish; shellfish; algae; seaweed; Salicornia; oysters; clams; abalone; shrimp; carnivores; herbivores; omnivores; detritivores; nutrients Abstract Israeli aquaculture began in the 1920s, with common carp monoculture. This was followed by polyculture of carp with tilapias, grey mullet, and planktivorous carp. Scientific research on polyculture started in the 1950s and has since contributed to the global science and practice of green water aquaculture, especially with novel polyculture approaches and concepts. Today, the industry is characterized by intensive freshwater polyculture, implemented in earthen fish ponds and reservoirs. In the Mediterranean coastal plain, fresh, brackish, and marine water polyculture is carried out in semi-intensive fishponds. Polyculture in Israel is an entrepreneurial activity that combines ecological principles of Chinese polyculture with local technologies and objectives. The Biofloc approach (active suspension ponds, ASP), periphyton, and aquaponics, were developed in the 1980s in response to rising public and policymakers‘ concerns and regulations on land use, pollution, use of chemicals, and organic manures. -

The IMPACT of HARMFUL ALGAL BLOOMS on FINFISH MORTALITY, PATHOLOGY and TOXICOLOGY
The IMPACT of HARMFUL ALGAL BLOOMS on FINFISH MORTALITY, PATHOLOGY and TOXICOLOGY "Et toutes les eaux qui étaient dans la rivière s'étaient changées en sang. Et les poissons se trouvaient dans la rivière et moururent. Et la rivière empestait, et les Egyptiens ne pouvaient pas boire l'eau de la rivière. " (Livre de l'Exode, 7, 20-24) REPERES OCÉAN N° 10-1995 The IMPACT of HARMFUL ALGAL BLOOMS on FINFISH MORTALITY, PATHOLOGY and TOXICOLOGY IMPACT des TOXINES ALGALES sur les POISSONS MORTALITÉ, PATHOLOGIE, TOXICOLOGIE Jacques BRUSLE Université de Perpignan, laboratoire de biologie marine REMER 6e CONFÉRENCE INTERNATIONALE SUR LE PHYTOPLANCTON MARIN Nantes, 18-22 octobre 1993 sous le titre "The impact of harmful algal blooms on finfish : occurrence of fish kills, pathology, toxicological mechanisms, ecological and economic impacts. A review." par Jacques BRUSLE Laboratoire de biologie marine, Université de Perpignan 52, avenue de Villeneuve, 66860 PERPIGNAN cedex - FRANCE EDITIONS IFREMER Centre de Brest B.P. 70 - 29280 PLOUZANÉ (France) Tél. 98 22 40 13 - Fax : 98 22 45 86 ISBN 2-905434-64-3 ISSN 1240-1153 Institut français de recherche pour l'exploitation de la mer, IFREMER, 1995 TABLE DES MATIÈRES SOMMAIRE EN FRANÇAIS 5 INTRODUCTION 9 CHAPTER I - OCCURRENCE OF FISH KILLS ASSOCIATED WITH "RED TIDES" 11 Europe 13 North america 14 Indo pacific region 14 Other countries 15 CHAPTER II - ALGAL SPECIES INVOLVED IN FISH KILLS 25 CHAPTER m - CAUSATIVE MECHANISMS OF FISH DEATH 35 02 depletion 35 Mechanical damage to gills 36 Chemical injury from -

Taxonomy and Physiology of Oxyrrhis Marina and Oxyrrhis Maritima in Korean Waters
water Article Taxonomy and Physiology of Oxyrrhis marina and Oxyrrhis maritima in Korean Waters Min Kyoung Jung 1, Tae Yeon Yin 1, Seung Joo Moon 2, Jaeyeon Park 2,* and Eun Young Yoon 1,* 1 Climate Change Research Laboratory, Advanced Institute of Convergence Technology, Suwon 16229, Korea; [email protected] (M.K.J.); [email protected] (T.Y.Y.) 2 Environment and Resource Convergence Center, Advanced Institute of Convergence Technology, Suwon 16229, Korea; [email protected] * Correspondence: [email protected] (J.P.); [email protected] (E.Y.Y.) Abstract: The genus Oxyrrhis is a heterotrophic dinoflagellate found in diverse marine environments. Oxyrrhis spp. have received attention owing to their ecological and industrial importance, high lipid contents, and docosahexaenoic acid formation. To the best of our knowledge, contrary to O. marina, ecophysiological characterization studies on O. maritima have not yet been reported. Therefore, we investigated the taxonomy and ecophysiology of four strains of O. marina from coastal waters and two strains of O. maritima from the littoral tidepool waters of Korea. Based on phylogenetic trees constructed using internal transcribed spacer ribosomal DNA (ITS rDNA) and SSU rDNA of dinoflagellates, the clade of all four O. marina strains was divergent from that of the two O. maritima strains. We measured the growth rates of both species at various water temperatures (10–36 ◦C), salinities (5–90), and light intensities (0–100 µE·m−2·s−1). The lowest (O. marina and O. maritima: ◦ ◦ ◦ 10 C) and highest temperatures (O. marina: <35 C, O. maritima: >35 C) revealed that O. -

Grazing Interactions Between Oxyrrhis Marina and Synechococcus Strains Grown in Single Nitrogen Sources
Western Washington University Western CEDAR WWU Graduate School Collection WWU Graduate and Undergraduate Scholarship 2010 Grazing interactions between Oxyrrhis marina and Synechococcus strains grown in single nitrogen sources Virginia. Selz Western Washington University Follow this and additional works at: https://cedar.wwu.edu/wwuet Part of the Marine Biology Commons Recommended Citation Selz, Virginia., "Grazing interactions between Oxyrrhis marina and Synechococcus strains grown in single nitrogen sources" (2010). WWU Graduate School Collection. 105. https://cedar.wwu.edu/wwuet/105 This Masters Thesis is brought to you for free and open access by the WWU Graduate and Undergraduate Scholarship at Western CEDAR. It has been accepted for inclusion in WWU Graduate School Collection by an authorized administrator of Western CEDAR. For more information, please contact [email protected]. Grazing Interactions between Oxyrrhis marina and Synechococcus Strains Grown in Single Nitrogen Sources By Virginia Selz Accepted in Partial Completion Of the Requirements for the Degree Master of Science ____________________________ Dr. Moheb A. Ghali, Dean of the Graduate School ADVISORY COMMITTEE ______________________________ Chair, Dr. Deborah Donovan ______________________________ Co-chair, Dr. Suzanne Strom _______________________________ Dr. Marion Brodhagen MASTER’S THESIS In presenting this thesis in partial fulfillment of the requirements for a master’s degree at Western Washington University, I grant to Western Washington University the non‐exclusive royalty‐free right to archive, reproduce, distribute, and display the thesis in any and all forms, including electronic format via any digital library mechanisms maintained by WWU. I represent and warrant this is my original work, and does not infringe or violate any rights of others.